The `DeeDeeExperiment` User's Guide
Najla Abassi
Institute of Medical Biostatistics, Epidemiology and Informatics (IMBEI), Mainzabassina@uni-mainz.de
Lea Rothörl
Institute of Medical Biostatistics, Epidemiology and Informatics (IMBEI), MainzFederico Marini
Institute of Medical Biostatistics, Epidemiology and Informatics (IMBEI), MainzResearch Center for Immunotherapy (FZI), Mainzmarinif@uni-mainz.de
1 July 2025
Source:vignettes/DeeDeeExperiment_manual.Rmd
DeeDeeExperiment_manual.RmdIntroduction
Why do we need a new class?
Differential expression analysis (DEA) and functional enrichment analysis (FEA) are core steps in transcriptomic workflows, helping researchers detect and interpret biological differences between conditions. However, as the number of conditions (defined by complex experimental designs) and analysis tools increases, managing the outputs of these analyses across multiple contrasts becomes increasingly overwhelming. This challenge is further amplified in single-cell RNA-seq, where pseudo-bulk analyses generate numerous results tables across cell types and contrasts, making it challenging to organize, explore, and reproduce findings, even for experienced users.
To address these issues, we introduce the DeeDeeExperiment
class, an S4 object that extends the widely adopted
SummarizedExperiment class in Bioconductor.
DeeDeeExperiment provides a structured and consistent
framework for integrating DEA and FEA results alongside core expression
data and metadata. By centralizing these results in a single container,
DeeDeeExperiment enables users to retrieve, explore, and
manage their analysis results more efficiently across multiple
contrasts.
This vignette introduces the class structure, demonstrates how to
create and work with DeeDeeExperiment objects, and walks
through a typical RNA-seq workflow using the class.
Anatomy of DeeDeeExperiment
The DeeDeeExperiment class inherits from the core
Bioconductor SummarizedExperiment object. This means it
inherits all the slots, methods, and compatibility with tools built for
SummarizedExperiment, while introducing additional
components to streamline downstream analysis.
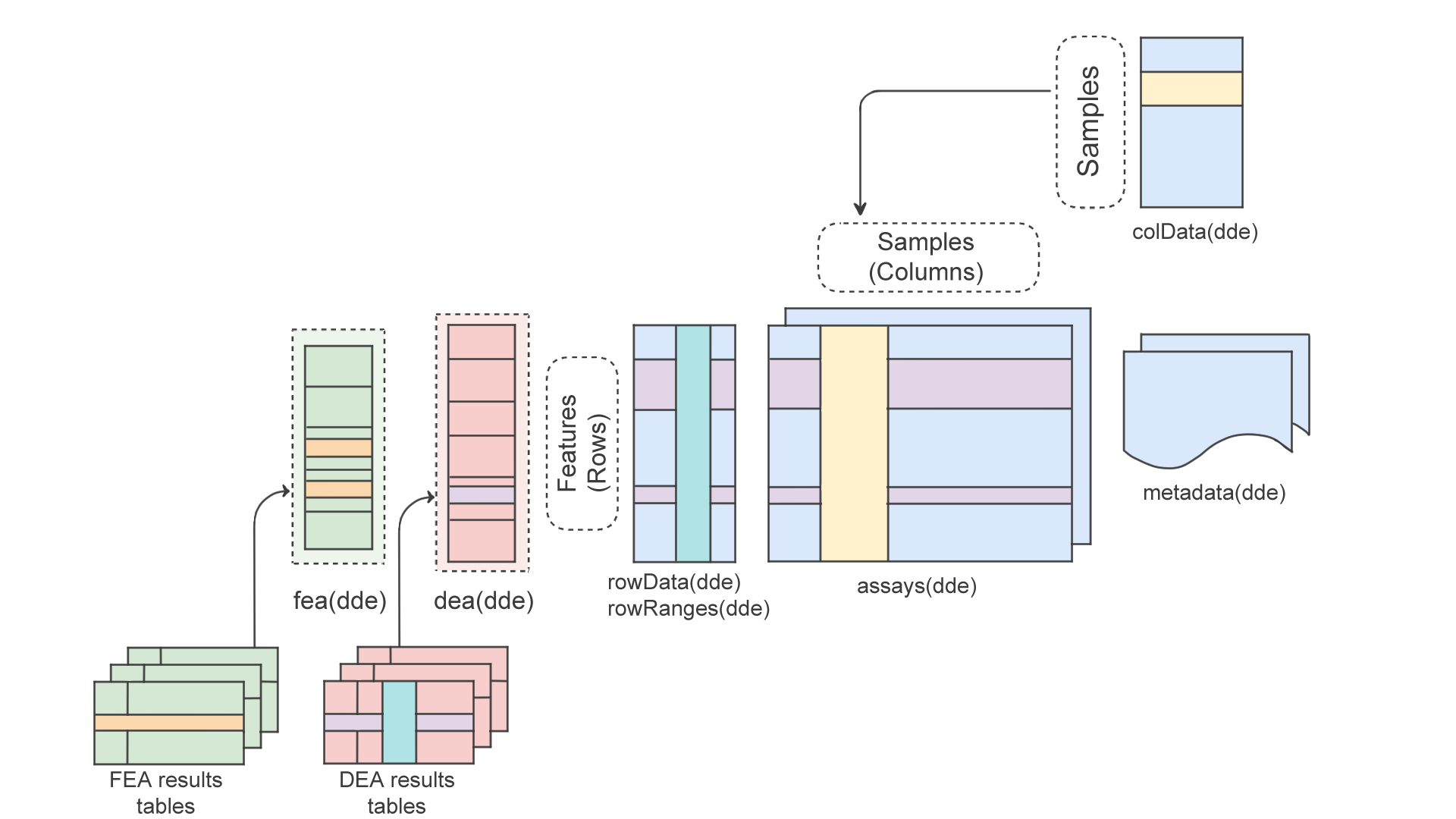
Specifically, DeeDeeExperiment has two new slots:
-
dea: a list that stores results from differential expression analysis (DEA), along with relevant metadata. This includes:
The main columns to use in DEA in addition to the feature identifier
(log2FoldChange, pvalue, and
padj) are also stored in the rowData of the
DeeDeeExperiment object.
-
fea: a list that stores results from functional enrichment analysis (FEA), along with relevant metadata. This includes:- The name of the associated DEA contrast.
- The FEA name.
- A
GeneTonicList-compatible set ofshaken_results(ready to be used in GeneTonic). - The enrichment tool used to generate the results
(e.g.
topGO,clusterProfiler...). - A copy of the original enrichment result object.
Creating a DeeDeeExperiment object
In order to create a DeeDeeExperiment object, at least
one of the following inputs are required:
-
se: ASummarizedExperimentobject, that will be used as a scaffold to store the DE related information. -
de_results: A named list of DE results, in any of the formats supported by the package (currently results fromDESeq2,edgeR, orlimma). -
enrich_results: A named list of FE results, asdata.framegenerated by one of the following packages (currentlytopGO,clusterProfiler,enrichR,gProfiler,fgsea,gsea,DAVID, and output ofGeneTonicshakers)
# do not run
dde <- DeeDeeExperiment(
se = se, # se is a SummarizedExperiment object
# de_results is a named list of de results
de_results = de_results,
# enrich_res is a named list of enrichment results
enrich_results = enrich_results
)Getting started
To install this package, start R and enter:
if (!requireNamespace("BiocManager", quietly = TRUE)) {
install.packages("BiocManager")
}
BiocManager::install("DeeDeeExperiment")Once installed, the package can be loaded and attached to your current workspace as follows:
DeeDeeExperiment on the macrophage
dataset
In the remainder of this vignette, we will illustrate the main features of DeeDeeExperiment on a publicly available dataset from Alasoo, et al. “Shared genetic effects on chromatin and gene expression indicate a role for enhancer priming in immune response”, published in Nature Genetics, January 2018 (Alasoo et al. 2018) doi:10.1038/s41588-018-0046-7.
The data is made available via the macrophage Bioconductor package, which contains the files output from the Salmon quantification (version 0.12.0, with GENCODE v29 reference), as well as the values summarized at the gene level, which we will use to exemplify.
In the macrophage experimental setting, the samples are
available from 6 different donors, in 4 different conditions (naive,
treated with Interferon gamma, with SL1344, or with a combination of
Interferon gamma and SL1344).
Let’s start by loading all the necessary packages:
library("DeeDeeExperiment")
library("macrophage")
library("DESeq2")
library("edgeR")
library("DEFormats")We will demonstrate how DeeDeeExperiment fits into a
regular bulk RNA-seq data analysis workflow. For this we will start by
processing the data and setting up the design for the Differential
Expression Analysis (DEA).
# load data
data(gse, package = "macrophage")If you already have your differential expression results and want to
skip the DEA workflow examples, you can jump directly to the section 4 Creating
DeeDeeExperiment the DeeDee way - otherwise, you can follow along in
the subsequent sections where we will showcase how the typical objects
from each DE framework will seamlessly fit into a
DeeDeeExperiment object.
The remainder of this vignette assumes some knowledge of Differential
Expression Analysis workflows. If you want some excellent introductions
to this topic, you can refer to (Love et al.
2016) and (Law et al. 2018) -
Otherwise, you can check (Ludt et al.
2022) for a comprehensive overview of these workflows, as well as
the usage of tools such as GeneTonic or
pcaExproler for interactive exploration.
DEA: DESeq2 framework
Below we will demonstrate an example on how to generate DE results
with DESeq2
# set up design
dds_macrophage <- DESeqDataSet(gse, design = ~ line + condition)
rownames(dds_macrophage) <- substr(rownames(dds_macrophage), 1, 15)
keep <- rowSums(counts(dds_macrophage) >= 10) >= 6
dds_macrophage <- dds_macrophage[keep, ]For the sake of demonstration, we will further subset the dataset for faster processing
# set seed for reproducibility
set.seed(42)
# sample randomly for 1k genes to speed up the processing
selected_genes <- sample(rownames(dds_macrophage), 1000)
dds_macrophage <- dds_macrophage[selected_genes, ]We then run the main DESeq() function and check the
resultsNames being generated
dds_macrophage
#> class: DESeqDataSet
#> dim: 1000 24
#> metadata(7): tximetaInfo quantInfo ... txdbInfo version
#> assays(3): counts abundance avgTxLength
#> rownames(1000): ENSG00000164741 ENSG00000078808 ... ENSG00000273680
#> ENSG00000073536
#> rowData names(2): gene_id SYMBOL
#> colnames(24): SAMEA103885102 SAMEA103885347 ... SAMEA103885308
#> SAMEA103884949
#> colData names(15): names sample_id ... condition line
# run DESeq
dds_macrophage <- DESeq(dds_macrophage)
# contrasts
resultsNames(dds_macrophage)
#> [1] "Intercept" "line_eiwy_1_vs_diku_1"
#> [3] "line_fikt_3_vs_diku_1" "line_ieki_2_vs_diku_1"
#> [5] "line_podx_1_vs_diku_1" "line_qaqx_1_vs_diku_1"
#> [7] "condition_IFNg_vs_naive" "condition_IFNg_SL1344_vs_naive"
#> [9] "condition_SL1344_vs_naive"Let’s extract the DE results for each contrast. We will only use two contrasts
FDR <- 0.05
# IFNg_vs_naive
IFNg_vs_naive <- results(dds_macrophage,
name = "condition_IFNg_vs_naive",
lfcThreshold = 1,
alpha = FDR
)
IFNg_vs_naive <- lfcShrink(dds_macrophage,
coef = "condition_IFNg_vs_naive",
res = IFNg_vs_naive,
type = "apeglm"
)
# Salm_vs_naive
Salm_vs_naive <- results(dds_macrophage,
name = "condition_SL1344_vs_naive",
lfcThreshold = 1,
alpha = FDR
)
Salm_vs_naive <- lfcShrink(dds_macrophage,
coef = "condition_SL1344_vs_naive",
res = Salm_vs_naive,
type = "apeglm"
)
head(IFNg_vs_naive)
#> log2 fold change (MAP): condition IFNg vs naive
#> Wald test p-value: condition IFNg vs naive
#> DataFrame with 6 rows and 5 columns
#> baseMean log2FoldChange lfcSE pvalue padj
#> <numeric> <numeric> <numeric> <numeric> <numeric>
#> ENSG00000164741 918.0347 0.2291487 0.3006751 9.80541e-01 1.00000e+00
#> ENSG00000078808 6684.4755 -0.0115336 0.0729923 1.00000e+00 1.00000e+00
#> ENSG00000251034 12.1241 -0.1167013 0.4049828 9.33899e-01 1.00000e+00
#> ENSG00000162676 10.2220 0.2691481 0.3776971 9.00817e-01 1.00000e+00
#> ENSG00000170356 15.0772 -0.1278637 0.3522078 9.72815e-01 1.00000e+00
#> ENSG00000204257 1922.0256 4.0550244 0.1960055 1.67298e-56 8.36489e-54
head(Salm_vs_naive)
#> log2 fold change (MAP): condition SL1344 vs naive
#> Wald test p-value: condition SL1344 vs naive
#> DataFrame with 6 rows and 5 columns
#> baseMean log2FoldChange lfcSE pvalue padj
#> <numeric> <numeric> <numeric> <numeric> <numeric>
#> ENSG00000164741 918.0347 0.0114691 0.3224207 0.999632 1.000000
#> ENSG00000078808 6684.4755 0.7893601 0.0734122 0.997559 1.000000
#> ENSG00000251034 12.1241 1.1881497 0.4894973 0.226918 0.982042
#> ENSG00000162676 10.2220 0.4420821 0.4228376 0.843212 1.000000
#> ENSG00000170356 15.0772 -0.6249201 0.4252786 0.726972 1.000000
#> ENSG00000204257 1922.0256 -0.7293810 0.2005145 0.883888 1.000000We now have two DESeqResults objects:
IFNg_vs_naive and Salm_vs_naive, ready to be
integrated into a DeeDeeExperiment object.
DEA: limma framework
Not working with DESeq2? No problem.
DeeDeeExperiment is method-agnostic and easily integrates
results from other frameworks like limma. In this section,
we will walk through a typical limma workflow to generate
DE results.
For demonstration purposes, we convert the filtered
dds_macrophage as input for limma, using the
as.DGEList() function from the DEFormats
package.
# create DGE list
dge <- DEFormats::as.DGEList(dds_macrophage)We then normalize and voom-transform the expression values, before
running the lmFit() function:
# normalize the counts
dge <- calcNormFactors(dge)
# create design for DE
design <- model.matrix(~ line + group, data = dge$samples)
# transform counts into logCPM
v <- voom(dge, design)
# fitting linear models using weighted least squares for each gene
fit <- lmFit(v, design)We define here as usual the contrast matrix:
# available comparisons
colnames(design)
#> [1] "(Intercept)" "lineeiwy_1" "linefikt_3" "lineieki_2"
#> [5] "linepodx_1" "lineqaqx_1" "groupIFNg" "groupIFNg_SL1344"
#> [9] "groupSL1344"
# setup comparisons
contrast_matrix <- makeContrasts(
IFNg_vs_Naive = groupIFNg,
Salm_vs_Naive = groupSL1344,
levels = design
)We then extract the results by applying the
contrasts.fit function, followed by the empirical Bayes
moderation as it follows:
# apply contrast
fit2 <- contrasts.fit(fit, contrast_matrix)
# empirical Bayes moderation of standard errors
fit2 <- eBayes(fit2)
de_limma <- fit2 # MArrayLM objectWe show here the top 10 DE genes, as detected in the contrast
IFNg_vs_Naive
# show top 10 genes, first columns
topTable(de_limma, coef = "IFNg_vs_Naive", number = 10)[, 1:7]
#> seqnames start end width strand gene_id
#> ENSG00000204257 chr6 32948613 32969094 20482 - ENSG00000204257.14
#> ENSG00000231389 chr6 33064569 33080775 16207 - ENSG00000231389.7
#> ENSG00000123992 chr2 219373527 219400022 26496 - ENSG00000123992.19
#> ENSG00000197448 chr7 143244093 143270854 26762 + ENSG00000197448.13
#> ENSG00000227531 chr9 111139246 111284836 145591 - ENSG00000227531.1
#> ENSG00000135124 chr12 121209857 121234106 24250 + ENSG00000135124.14
#> ENSG00000075399 chr16 89707134 89720986 13853 - ENSG00000075399.13
#> ENSG00000065911 chr2 74198562 74217565 19004 + ENSG00000065911.11
#> ENSG00000233621 chr1 37454879 37474411 19533 - ENSG00000233621.1
#> ENSG00000104894 chr19 49335171 49343335 8165 + ENSG00000104894.11
#> SYMBOL
#> ENSG00000204257 HLA-DMA
#> ENSG00000231389 HLA-DPA1
#> ENSG00000123992 DNPEP
#> ENSG00000197448 GSTK1
#> ENSG00000227531 <NA>
#> ENSG00000135124 P2RX4
#> ENSG00000075399 VPS9D1
#> ENSG00000065911 MTHFD2
#> ENSG00000233621 LINC01137
#> ENSG00000104894 CD37We now have a MArrayLM object: de_limma,
ready to be integrated into a DeeDeeExperiment object.
DEA: edgeR framework
Below we will walk you through a typical edgeR pipeline
to produce DE results ready for integration in
DeeDeeExperiment.
We reuse the normalized DGEList and design matrix from
the previous limma workflow (again, created via
DEFormats::as.DGEList()):
dge <- DEFormats::as.DGEList(dds_macrophage)
# estimate dispersion
dge <- estimateDisp(dge, design)
# perform likelihood ratio test
fit <- glmFit(dge, design)Here as well, we define the contrasts of interest:
# available comparisons
colnames(design)
#> [1] "(Intercept)" "lineeiwy_1" "linefikt_3" "lineieki_2"
#> [5] "linepodx_1" "lineqaqx_1" "groupIFNg" "groupIFNg_SL1344"
#> [9] "groupSL1344"
# setup comparisons
contrast_matrix <- makeContrasts(
IFNg_vs_Naive = groupIFNg,
Salm_vs_Naive = groupSL1344,
levels = design
)… and. we create the DGELRT objects to store the DE
results as follows:
# DGELRT objects
dge_lrt_IFNg_vs_naive <- glmLRT(fit,
contrast = contrast_matrix[, "IFNg_vs_Naive"])
dge_lrt_Salm_vs_naive <- glmLRT(fit,
contrast = contrast_matrix[, "Salm_vs_Naive"])The top 10 DE genes computed within the edgeR framework can be shown as in this chunk:
# show top 10 genes, first few columns
topTags(dge_lrt_IFNg_vs_naive, n = 10)[, 1:7]
#> Coefficient: 1*groupIFNg
#> seqnames start end width strand gene_id
#> ENSG00000204257 chr6 32948613 32969094 20482 - ENSG00000204257.14
#> ENSG00000231389 chr6 33064569 33080775 16207 - ENSG00000231389.7
#> ENSG00000123992 chr2 219373527 219400022 26496 - ENSG00000123992.19
#> ENSG00000197448 chr7 143244093 143270854 26762 + ENSG00000197448.13
#> ENSG00000227531 chr9 111139246 111284836 145591 - ENSG00000227531.1
#> ENSG00000233621 chr1 37454879 37474411 19533 - ENSG00000233621.1
#> ENSG00000065911 chr2 74198562 74217565 19004 + ENSG00000065911.11
#> ENSG00000135124 chr12 121209857 121234106 24250 + ENSG00000135124.14
#> ENSG00000163131 chr1 150730196 150765957 35762 - ENSG00000163131.10
#> ENSG00000100418 chr22 41598028 41621096 23069 - ENSG00000100418.7
#> SYMBOL
#> ENSG00000204257 HLA-DMA
#> ENSG00000231389 HLA-DPA1
#> ENSG00000123992 DNPEP
#> ENSG00000197448 GSTK1
#> ENSG00000227531 <NA>
#> ENSG00000233621 LINC01137
#> ENSG00000065911 MTHFD2
#> ENSG00000135124 P2RX4
#> ENSG00000163131 CTSS
#> ENSG00000100418 DESI1For the sake of demonstrating that DeeDeeExperiment can
handle the different objects from the edgeR framework, we
also compute some results with the exactTest()
approach.
# perform exact test
# exact test doesn't handle multi factor models, so we have to subset
# IFNg vs naive
keep_samples <- dge$samples$group %in% c("naive", "IFNg")
dge_sub <- dge[, keep_samples]
# droplevel
group <- droplevels(dge_sub$samples$group)
dge_sub$samples$group <- group
# recalculate normalization and dispersion
dge_sub <- calcNormFactors(dge_sub)
dge_sub <- estimateDisp(dge_sub, design = model.matrix(~group))
# exact test
# DGEExact object
dge_exact_IFNg_vs_naive <- exactTest(dge_sub, pair = c("naive", "IFNg"))
# SL1344 vs naive
keep_samples <- dge$samples$group %in% c("naive", "SL1344")
dge_sub <- dge[, keep_samples]
# droplevel
group <- droplevels(dge_sub$samples$group)
dge_sub$samples$group <- group
# recalculate normalization and dispersion
dge_sub <- calcNormFactors(dge_sub)
dge_sub <- estimateDisp(dge_sub, design = model.matrix(~group))
# exact test
dge_exact_Salm_vs_naive <- exactTest(dge_sub, pair = c("naive", "SL1344"))Again, the top 10 DE genes in the
dge_exact_Salm_vs_naive object can be shown with
topTags(dge_exact_Salm_vs_naive, n = 10)[, 1:7]
#> Comparison of groups: SL1344-naive
#> seqnames start end width strand gene_id
#> ENSG00000234394 chr9 68306528 68330626 24099 + ENSG00000234394.8
#> ENSG00000132405 chr4 6909242 7033118 123877 + ENSG00000132405.18
#> ENSG00000109756 chr4 159103013 159360174 257162 + ENSG00000109756.9
#> ENSG00000078804 chr20 34704290 34713439 9150 + ENSG00000078804.12
#> ENSG00000105711 chr19 35030466 35040449 9984 + ENSG00000105711.11
#> ENSG00000188211 chr11 17351726 17377341 25616 + ENSG00000188211.8
#> ENSG00000214113 chr6 5102593 5260939 158347 - ENSG00000214113.10
#> ENSG00000253535 chr8 24295814 24912073 616260 - ENSG00000253535.5
#> ENSG00000102780 chr13 42040036 42256578 216543 + ENSG00000102780.16
#> ENSG00000179826 chr11 18120955 18138480 17526 + ENSG00000179826.6
#> SYMBOL
#> ENSG00000234394 <NA>
#> ENSG00000132405 TBC1D14
#> ENSG00000109756 RAPGEF2
#> ENSG00000078804 TP53INP2
#> ENSG00000105711 SCN1B
#> ENSG00000188211 NCR3LG1
#> ENSG00000214113 LYRM4
#> ENSG00000253535 <NA>
#> ENSG00000102780 DGKH
#> ENSG00000179826 MRGPRX3We now have two DGELRT & two DGEExact
objects: dge_lrt_IFNg_vs_naive,
dge_lrt_Salm_vs_naive &
dge_exact_IFNg_vs_naive,
dge_exact_Salm_vs_naive respectively, ready to be
integrated into a DeeDeeExperiment object.
For demonstration purposes, we will use precomputed FEA results
generated with run_topGO() (a practical wrapper provided by
the mosdef package), and with the gost()
function from gprofiler2 package.
# load Functional Enrichment Analysis results examples
data("topGO_results_list", package = "DeeDeeExperiment")
data("gost_res", package = "DeeDeeExperiment")
data("clusterPro_res", package = "DeeDeeExperiment")The enrichment results can be displayed in a summary way with these commands:
# ifng vs naive
head(topGO_results_list$ifng_vs_naive)
#> GO.ID
#> 1 GO:0002822
#> 2 GO:0002697
#> 3 GO:0002250
#> 4 GO:1904064
#> 5 GO:0042098
#> 6 GO:0070663
#> Term
#> 1 regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
#> 2 regulation of immune effector process
#> 3 adaptive immune response
#> 4 positive regulation of cation transmembrane transport
#> 5 T cell proliferation
#> 6 regulation of leukocyte proliferation
#> Annotated Significant Expected Rank in p.value_classic p.value_elim
#> 1 11 6 0.55 2 3.90e-06
#> 2 13 6 0.65 5 1.30e-05
#> 3 17 10 0.85 1 3.70e-05
#> 4 10 4 0.50 13 8.80e-04
#> 5 10 4 0.50 14 8.80e-04
#> 6 11 4 0.55 17 1.34e-03
#> p.value_classic
#> 1 3.90e-06
#> 2 1.30e-05
#> 3 4.00e-10
#> 4 8.80e-04
#> 5 8.80e-04
#> 6 1.34e-03
#> genes
#> 1 CD28,CD69,CR1L,IL27,SECTM1,TNFSF18
#> 2 CD28,CD69,CR1L,IL27,SECTM1,TNFSF18
#> 3 CD28,CD69,CR1L,CTSS,HLA-DMA,HLA-DPA1,IL27,LAMP3,SECTM1,TNFSF18
#> 4 ANK2,CTSS,KCNE5,P2RX4
#> 5 CD28,HLA-DPA1,IL27,TNFSF18
#> 6 CD28,HLA-DPA1,IL27,TNFSF18
# salmonella vs naive
head(gost_res$result)
#> query significant p_value term_size query_size intersection_size
#> 1 query_1 TRUE 0.01425000 2551 85 30
#> 2 query_1 TRUE 0.03856196 625 85 13
#> 3 query_1 TRUE 0.03856196 625 85 13
#> 4 query_1 TRUE 0.03904673 1616 85 22
#> 5 query_1 FALSE 0.39973004 164 85 6
#> 6 query_1 FALSE 0.41892532 791 85 13
#> precision recall term_id source
#> 1 0.35294118 0.01176009 GO:0042221 GO:BP
#> 2 0.15294118 0.02080000 GO:0034097 GO:BP
#> 3 0.15294118 0.02080000 GO:1901652 GO:BP
#> 4 0.25882353 0.01361386 GO:0070887 GO:BP
#> 5 0.07058824 0.03658537 GO:0071356 GO:BP
#> 6 0.15294118 0.01643489 GO:1901701 GO:BP
#> term_name effective_domain_size
#> 1 response to chemical 16175
#> 2 response to cytokine 16175
#> 3 response to peptide 16175
#> 4 cellular response to chemical stimulus 16175
#> 5 cellular response to tumor necrosis factor 16175
#> 6 cellular response to oxygen-containing compound 16175
#> source_order parents
#> 1 10049 GO:0050896
#> 2 8408 GO:1901652
#> 3 21619 GO:0042221
#> 4 16358 GO:00422....
#> 5 16608 GO:00346....
#> 6 21654 GO:00708....
#> evidence_codes
#> 1 IMP,IDA,IDA IBA,IDA IMP ISS,IDA ISS IBA,IDA TAS,IMP,TAS,IDA,IBA,ISS,ISS IBA,IDA IMP IGI IBA,IDA,IMP,IDA ISS,IDA,IEP IBA,IEP,IDA IEP,IDA IEP,IMP,IBA,ISS,IDA IEP ISS IBA TAS,IDA,IMP IBA,ISS,IDA,ISS
#> 2 IMP,IDA IBA,ISS,ISS,ISS IBA,IDA IMP IGI IBA,IDA,IDA,IEP,IDA,IDA IEP ISS IBA,IDA,ISS
#> 3 IMP,IDA IBA,ISS,ISS,ISS IBA,IDA IMP IGI IBA,IDA,IDA,IEP,IDA,IDA IEP ISS IBA,IDA,ISS
#> 4 IDA,IDA IMP ISS,IDA IBA,IDA TAS,IMP,TAS,IDA,IBA,ISS,IDA IMP IBA,IMP,ISS,IDA,IEP IBA,IDA IEP,IDA,ISS,IDA ISS IBA,IDA,IMP IBA,ISS,ISS
#> 5 ISS IBA,IDA IMP IGI,IDA,IEP,IEP ISS IBA,ISS
#> 6 IDA,IDA,IDA IBA,IMP,TAS,IDA,IDA IMP IBA,IMP,IDA,ISS,IDA,IMP,ISS
#> intersection
#> 1 LAMP3,RAPGEF2,IL12RB2,P2RX4,SHPK,CYP3A5,ADPRS,AGRN,TRERF1,SRXN1,UGCG,TNFSF18,RELA,NAIP,OSBPL7,CH25H,DNAJA1,MT1A,HAS2,PDE2A,SLC11A2,HAMP,KHK,GREM1,CCL2,LCOR,TSHR,GPR37,ADAR,OCSTAMP
#> 2 LAMP3,IL12RB2,SHPK,UGCG,TNFSF18,RELA,NAIP,CH25H,HAS2,PDE2A,CCL2,ADAR,OCSTAMP
#> 3 LAMP3,IL12RB2,SHPK,UGCG,TNFSF18,RELA,NAIP,CH25H,HAS2,PDE2A,CCL2,ADAR,OCSTAMP
#> 4 RAPGEF2,P2RX4,SHPK,CYP3A5,ADPRS,AGRN,TRERF1,SRXN1,UGCG,RELA,OSBPL7,CH25H,DNAJA1,MT1A,PDE2A,SLC11A2,GREM1,CCL2,LCOR,TSHR,GPR37,OCSTAMP
#> 5 TNFSF18,RELA,NAIP,HAS2,CCL2,OCSTAMP
#> 6 RAPGEF2,P2RX4,SHPK,ADPRS,AGRN,TRERF1,RELA,OSBPL7,PDE2A,CCL2,LCOR,TSHR,GPR37
# ifng vs naive
head(clusterPro_res$ifng_vs_naive)
#> ID
#> GO:0002250 GO:0002250
#> GO:0002819 GO:0002819
#> GO:0002822 GO:0002822
#> GO:0002460 GO:0002460
#> GO:0002697 GO:0002697
#> GO:0050776 GO:0050776
#> Description
#> GO:0002250 adaptive immune response
#> GO:0002819 regulation of adaptive immune response
#> GO:0002822 regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
#> GO:0002460 adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
#> GO:0002697 regulation of immune effector process
#> GO:0050776 regulation of immune response
#> GeneRatio BgRatio RichFactor FoldEnrichment zScore pvalue
#> GO:0002250 10/37 17/744 0.5882353 11.828299 10.325309 3.958927e-10
#> GO:0002819 6/37 11/744 0.5454545 10.968059 7.614485 3.872646e-06
#> GO:0002822 6/37 11/744 0.5454545 10.968059 7.614485 3.872646e-06
#> GO:0002460 6/37 12/744 0.5000000 10.054054 7.228759 7.465887e-06
#> GO:0002697 6/37 13/744 0.4615385 9.280665 6.885949 1.336486e-05
#> GO:0050776 9/37 41/744 0.2195122 4.413975 5.141149 7.666847e-05
#> p.adjust qvalue
#> GO:0002250 2.561426e-07 2.233668e-07
#> GO:0002819 8.352007e-04 7.283293e-04
#> GO:0002822 8.352007e-04 7.283293e-04
#> GO:0002460 1.207607e-03 1.053083e-03
#> GO:0002697 1.729413e-03 1.508119e-03
#> GO:0050776 8.267416e-03 7.209526e-03
#> geneID
#> GO:0002250 ENSG00000231389/ENSG00000204257/ENSG00000078081/ENSG00000141574/ENSG00000197721/ENSG00000178562/ENSG00000197272/ENSG00000120337/ENSG00000163131/ENSG00000110848
#> GO:0002819 ENSG00000141574/ENSG00000197721/ENSG00000178562/ENSG00000197272/ENSG00000120337/ENSG00000110848
#> GO:0002822 ENSG00000141574/ENSG00000197721/ENSG00000178562/ENSG00000197272/ENSG00000120337/ENSG00000110848
#> GO:0002460 ENSG00000141574/ENSG00000197721/ENSG00000178562/ENSG00000197272/ENSG00000120337/ENSG00000110848
#> GO:0002697 ENSG00000141574/ENSG00000197721/ENSG00000178562/ENSG00000197272/ENSG00000120337/ENSG00000110848
#> GO:0050776 ENSG00000231389/ENSG00000204257/ENSG00000141574/ENSG00000197721/ENSG00000178562/ENSG00000197272/ENSG00000120337/ENSG00000163131/ENSG00000110848
#> Count
#> GO:0002250 10
#> GO:0002819 6
#> GO:0002822 6
#> GO:0002460 6
#> GO:0002697 6
#> GO:0050776 9To check which formats of Functional Enrichment Analysis (FEA)
results are supported natively within DeeDeeExperiment,
simply run the following command:
DeeDeeExperiment::supported_fea_formats()
#> Format Package
#> 1 data.frame topGO
#> 2 enrichResult clusterProfiler
#> 3 gseaResult clusterProfiler
#> 4 fgseaResult fgsea
#> 5 data.frame gprofiler2
#> 6 data.frame enrichR
#> 7 data.frame DAVID
#> 8 data.frame GeneTonicBy now, we collected DEA results from three different frameworks and generated multiple FEA results. Each result has its own structure, columns, metadata, …
Managing and exploring this growing stack of outputs can quickly
become overwhelming. So let’s now see how DeeDeeExperiment
brings all these pieces together.
Creating DeeDeeExperiment the DeeDee way
In the following example we construct the dde object using a
dds object and named list of DE
results
# initialize DeeDeeExperiment with dds object and DESeq results (as a named list)
dde <- DeeDeeExperiment(
se = dds_macrophage,
de_results = list(
IFNg_vs_naive = IFNg_vs_naive,
Salm_vs_naive = Salm_vs_naive
)
)
dde
#> class: DeeDeeExperiment
#> dim: 1000 24
#> metadata(7): tximetaInfo quantInfo ... txdbInfo version
#> assays(7): counts abundance ... H cooks
#> rownames(1000): ENSG00000164741 ENSG00000078808 ... ENSG00000273680
#> ENSG00000073536
#> rowData names(58): gene_id SYMBOL ... Salm_vs_naive_pvalue
#> Salm_vs_naive_padj
#> colnames(24): SAMEA103885102 SAMEA103885347 ... SAMEA103885308
#> SAMEA103884949
#> colData names(15): names sample_id ... condition line
#> dea(2): IFNg_vs_naive, Salm_vs_naive
#> fea(0):You can see from the summary overview that the information on the two
dea has been added to the original object.
Adding results to a dde object
Adding DEA results
Additional DE results can be added using the add_dea()
method. These results are stored in both the dea slot and
in the rowData.
We can also add results as individual element of class
DESeqResults, MArrayLM, DGEExact
or DGELRT.
# add a named list of results from edgeR
dde <- add_dea(dde, dea = list(
dge_exact_IFNg_vs_naive = dge_exact_IFNg_vs_naive,
dge_lrt_IFNg_vs_naive = dge_lrt_IFNg_vs_naive
))
# add results from limma as a MArrayLM object
dde <- add_dea(dde, dea = de_limma)
dde
#> class: DeeDeeExperiment
#> dim: 1000 24
#> metadata(7): tximetaInfo quantInfo ... txdbInfo version
#> assays(7): counts abundance ... H cooks
#> rownames(1000): ENSG00000164741 ENSG00000078808 ... ENSG00000273680
#> ENSG00000073536
#> rowData names(67): gene_id SYMBOL ... de_limma_pvalue de_limma_padj
#> colnames(24): SAMEA103885102 SAMEA103885347 ... SAMEA103885308
#> SAMEA103884949
#> colData names(15): names sample_id ... condition line
#> dea(5): IFNg_vs_naive, Salm_vs_naive, dge_exact_IFNg_vs_naive, dge_lrt_IFNg_vs_naive, de_limma
#> fea(0):
# inspect the columns of the rowData
names(rowData(dde))
#> [1] "gene_id"
#> [2] "SYMBOL"
#> [3] "baseMean"
#> [4] "baseVar"
#> [5] "allZero"
#> [6] "dispGeneEst"
#> [7] "dispGeneIter"
#> [8] "dispFit"
#> [9] "dispersion"
#> [10] "dispIter"
#> [11] "dispOutlier"
#> [12] "dispMAP"
#> [13] "Intercept"
#> [14] "line_eiwy_1_vs_diku_1"
#> [15] "line_fikt_3_vs_diku_1"
#> [16] "line_ieki_2_vs_diku_1"
#> [17] "line_podx_1_vs_diku_1"
#> [18] "line_qaqx_1_vs_diku_1"
#> [19] "condition_IFNg_vs_naive"
#> [20] "condition_IFNg_SL1344_vs_naive"
#> [21] "condition_SL1344_vs_naive"
#> [22] "SE_Intercept"
#> [23] "SE_line_eiwy_1_vs_diku_1"
#> [24] "SE_line_fikt_3_vs_diku_1"
#> [25] "SE_line_ieki_2_vs_diku_1"
#> [26] "SE_line_podx_1_vs_diku_1"
#> [27] "SE_line_qaqx_1_vs_diku_1"
#> [28] "SE_condition_IFNg_vs_naive"
#> [29] "SE_condition_IFNg_SL1344_vs_naive"
#> [30] "SE_condition_SL1344_vs_naive"
#> [31] "WaldStatistic_Intercept"
#> [32] "WaldStatistic_line_eiwy_1_vs_diku_1"
#> [33] "WaldStatistic_line_fikt_3_vs_diku_1"
#> [34] "WaldStatistic_line_ieki_2_vs_diku_1"
#> [35] "WaldStatistic_line_podx_1_vs_diku_1"
#> [36] "WaldStatistic_line_qaqx_1_vs_diku_1"
#> [37] "WaldStatistic_condition_IFNg_vs_naive"
#> [38] "WaldStatistic_condition_IFNg_SL1344_vs_naive"
#> [39] "WaldStatistic_condition_SL1344_vs_naive"
#> [40] "WaldPvalue_Intercept"
#> [41] "WaldPvalue_line_eiwy_1_vs_diku_1"
#> [42] "WaldPvalue_line_fikt_3_vs_diku_1"
#> [43] "WaldPvalue_line_ieki_2_vs_diku_1"
#> [44] "WaldPvalue_line_podx_1_vs_diku_1"
#> [45] "WaldPvalue_line_qaqx_1_vs_diku_1"
#> [46] "WaldPvalue_condition_IFNg_vs_naive"
#> [47] "WaldPvalue_condition_IFNg_SL1344_vs_naive"
#> [48] "WaldPvalue_condition_SL1344_vs_naive"
#> [49] "betaConv"
#> [50] "betaIter"
#> [51] "deviance"
#> [52] "maxCooks"
#> [53] "IFNg_vs_naive_log2FoldChange"
#> [54] "IFNg_vs_naive_pvalue"
#> [55] "IFNg_vs_naive_padj"
#> [56] "Salm_vs_naive_log2FoldChange"
#> [57] "Salm_vs_naive_pvalue"
#> [58] "Salm_vs_naive_padj"
#> [59] "dge_exact_IFNg_vs_naive_log2FoldChange"
#> [60] "dge_exact_IFNg_vs_naive_pvalue"
#> [61] "dge_exact_IFNg_vs_naive_padj"
#> [62] "dge_lrt_IFNg_vs_naive_log2FoldChange"
#> [63] "dge_lrt_IFNg_vs_naive_pvalue"
#> [64] "dge_lrt_IFNg_vs_naive_padj"
#> [65] "de_limma_log2FoldChange"
#> [66] "de_limma_pvalue"
#> [67] "de_limma_padj"The dde object now contains 5 DEAs in its
dea slot. The rowData also includes a record
of the log2FoldChange, pvalue, and
padj values for each contrast.
It is possible to overwrite the results, when the argument
force is set to TRUE
dde <- add_dea(dde, dea = list(same_contrast = dge_lrt_Salm_vs_naive))
# overwrite results with the same name
# e.g. if the content of the same object has changed
dde <- add_dea(dde,
dea = list(same_contrast = dge_exact_Salm_vs_naive),
force = TRUE)
dde
#> class: DeeDeeExperiment
#> dim: 1000 24
#> metadata(7): tximetaInfo quantInfo ... txdbInfo version
#> assays(7): counts abundance ... H cooks
#> rownames(1000): ENSG00000164741 ENSG00000078808 ... ENSG00000273680
#> ENSG00000073536
#> rowData names(70): gene_id SYMBOL ... same_contrast_pvalue
#> same_contrast_padj
#> colnames(24): SAMEA103885102 SAMEA103885347 ... SAMEA103885308
#> SAMEA103884949
#> colData names(15): names sample_id ... condition line
#> dea(6): IFNg_vs_naive, Salm_vs_naive, dge_exact_IFNg_vs_naive, dge_lrt_IFNg_vs_naive, de_limma, same_contrast
#> fea(0):Adding FEA results
FEA results can be added to a dde object using the
add_fea() method. These results are stored in the
fea slot.
If the fea_tool argument was not specified, the internal
function used to generate the standardized format of FEA result is
automatically detected - this should work in most cases, but it can be
beneficial to explicitly specify the fea_tool used, also
for simple bookkeeping reasons.
# add FEA results as a named list
dde <- add_fea(dde,
fea = list(IFNg_vs_naive = topGO_results_list$ifng_vs_naive))
# add FEA results as a single object
dde <- add_fea(dde, fea = gost_res$result)
# add FEA results and specify the FEA tool
dde <- add_fea(dde, fea = clusterPro_res, fea_tool = "clusterProfiler")
dde
#> class: DeeDeeExperiment
#> dim: 1000 24
#> metadata(7): tximetaInfo quantInfo ... txdbInfo version
#> assays(7): counts abundance ... H cooks
#> rownames(1000): ENSG00000164741 ENSG00000078808 ... ENSG00000273680
#> ENSG00000073536
#> rowData names(70): gene_id SYMBOL ... same_contrast_pvalue
#> same_contrast_padj
#> colnames(24): SAMEA103885102 SAMEA103885347 ... SAMEA103885308
#> SAMEA103884949
#> colData names(15): names sample_id ... condition line
#> dea(6): IFNg_vs_naive, Salm_vs_naive, dge_exact_IFNg_vs_naive, dge_lrt_IFNg_vs_naive, de_limma, same_contrast
#> fea(4): IFNg_vs_naive, gost_res$result, ifng_vs_naive, salmonella_vs_naiveLinking DE and FE Analysis in a dde object
Once DE and FE results have been added, FEAs can be linked to
specific DE contrasts using the link_dea_and_fea() method.
This maintains a clear association between the analyses and helps
document it within the dde object.
dde <- link_dea_and_fea(dde,
dea_name = "Salm_vs_naive",
fea_name = c("salmonella_vs_naive", "gost_res$result")
)This is then directly visible in the object summary, provided with
the dedicated summary() method:
summary(dde)
#> DE Results Summary:
#> DEA_name Up Down FDR
#> IFNg_vs_naive 36 17 0.05
#> Salm_vs_naive 90 34 0.05
#> dge_exact_IFNg_vs_naive 91 67 0.05
#> dge_lrt_IFNg_vs_naive 155 145 0.05
#> de_limma 265 303 0.05
#> same_contrast 168 187 0.05
#>
#> FE Results Summary:
#> FEA_Name Linked_DE FE_Type Term_Number
#> IFNg_vs_naive IFNg_vs_naive topGO 955
#> gost_res$result Salm_vs_naive gProfiler 2095
#> ifng_vs_naive . clusterProfiler 20
#> salmonella_vs_naive Salm_vs_naive clusterProfiler 11Adding contextual information
It is possible to add context to specific DEA results that are stored
in a dde object using the add_scenario_info()
method. This information can help document the experimental setup,
clarify comparisons, or maybe provided as additional element to a large
language model that can benefit of the extra bits to assist in the
interpretation.
dde <- add_scenario_info(dde,
dea_name = "IFNg_vs_naive",
info = "This results contains the output of a Differential Expression Analysis performed on data from the `macrophage` package, more precisely contrasting the counts from naive macrophage to those associated with IFNg."
)Summary of a dde object
All results stored in a dde object can be easily
inspected with the summary() method
# minimal summary
summary(dde)
#> DE Results Summary:
#> DEA_name Up Down FDR
#> IFNg_vs_naive 36 17 0.05
#> Salm_vs_naive 90 34 0.05
#> dge_exact_IFNg_vs_naive 91 67 0.05
#> dge_lrt_IFNg_vs_naive 155 145 0.05
#> de_limma 265 303 0.05
#> same_contrast 168 187 0.05
#>
#> FE Results Summary:
#> FEA_Name Linked_DE FE_Type Term_Number
#> IFNg_vs_naive IFNg_vs_naive topGO 955
#> gost_res$result Salm_vs_naive gProfiler 2095
#> ifng_vs_naive . clusterProfiler 20
#> salmonella_vs_naive Salm_vs_naive clusterProfiler 11It is possible to customize the output of summary()
using optional arguments. For example, the FDR threshold
can be adjusted to display the number of up- and downregulated genes for
each comparison.
# specify FDR threshold for subsetting DE genes based on adjusted p-values
summary(dde, FDR = 0.01)
#> DE Results Summary:
#> DEA_name Up Down FDR
#> IFNg_vs_naive 30 15 0.01
#> Salm_vs_naive 80 29 0.01
#> dge_exact_IFNg_vs_naive 69 41 0.01
#> dge_lrt_IFNg_vs_naive 124 108 0.01
#> de_limma 203 244 0.01
#> same_contrast 129 124 0.01
#>
#> FE Results Summary:
#> FEA_Name Linked_DE FE_Type Term_Number
#> IFNg_vs_naive IFNg_vs_naive topGO 955
#> gost_res$result Salm_vs_naive gProfiler 2095
#> ifng_vs_naive . clusterProfiler 20
#> salmonella_vs_naive Salm_vs_naive clusterProfiler 11It is also possible to include any scenario information associated
with each DEA by setting show_scenario_info = TRUE
# show contextual information, if available
summary(dde, show_scenario_info = TRUE)
#> DE Results Summary:
#> DEA_name Up Down FDR
#> IFNg_vs_naive 36 17 0.05
#> Salm_vs_naive 90 34 0.05
#> dge_exact_IFNg_vs_naive 91 67 0.05
#> dge_lrt_IFNg_vs_naive 155 145 0.05
#> de_limma 265 303 0.05
#> same_contrast 168 187 0.05
#>
#> FE Results Summary:
#> FEA_Name Linked_DE FE_Type Term_Number
#> IFNg_vs_naive IFNg_vs_naive topGO 955
#> gost_res$result Salm_vs_naive gProfiler 2095
#> ifng_vs_naive . clusterProfiler 20
#> salmonella_vs_naive Salm_vs_naive clusterProfiler 11
#>
#> Scenario Info:
#> - IFNg_vs_naive :
#> This results contains the output of a Differential Expression Analysis
#> performed on data from the `macrophage` package, more precisely contrasting
#> the counts from naive macrophage to those associated with IFNg.
#>
#>
#> No scenario info for: Salm_vs_naive, dge_exact_IFNg_vs_naive, dge_lrt_IFNg_vs_naive, de_limma, same_contrastRenaming results in a dde object
Specific results can be renamed after being added using the
rename_dea() and rename_fea() methods. The
corresponding column names in the rowData will be updated
accordingly.
# rename dea, one element
dde <- rename_dea(dde,
old_name = "de_limma",
new_name = "ifng_vs_naive_&_salm_vs_naive"
)
# multiple entries at once
dde <- rename_dea(dde,
old_name = c("dge_exact_IFNg_vs_naive", "dge_lrt_IFNg_vs_naive"),
new_name = c("exact_IFNg_vs_naive", "lrt_IFNg_vs_naive")
)
# rename fea
dde <- rename_fea(dde,
old_name = "gost_res$result",
new_name = "salm_vs_naive"
)Removing results in a dde object
The method remove_dea() and remove_fea()
can be used to delete results from a dde object.
# removing dea
dde <- remove_dea(dde, c(
"ifng_vs_naive_&_salm_vs_naive",
"exact_IFNg_vs_naive",
"dge_Salm_vs_naive"
))
# removing fea
dde <- remove_fea(dde, c("salmonella_vs_naive"))
dde
#> class: DeeDeeExperiment
#> dim: 1000 24
#> metadata(7): tximetaInfo quantInfo ... txdbInfo version
#> assays(7): counts abundance ... H cooks
#> rownames(1000): ENSG00000164741 ENSG00000078808 ... ENSG00000273680
#> ENSG00000073536
#> rowData names(64): gene_id SYMBOL ... same_contrast_pvalue
#> same_contrast_padj
#> colnames(24): SAMEA103885102 SAMEA103885347 ... SAMEA103885308
#> SAMEA103884949
#> colData names(15): names sample_id ... condition line
#> dea(4): IFNg_vs_naive, Salm_vs_naive, lrt_IFNg_vs_naive, same_contrast
#> fea(3): IFNg_vs_naive, salm_vs_naive, ifng_vs_naiveAccessing results stored in a dde object
Several accessor methods are available to retrieve DEA and FEA
results stored within a dde object.
dea_names() and fea_names() are the methods
to quickly retrieve the names of available DEAs and FEAs
# get DEA names
dea_names(dde)
#> [1] "IFNg_vs_naive" "Salm_vs_naive" "lrt_IFNg_vs_naive"
#> [4] "same_contrast"
# get FEA names
fea_names(dde)
#> [1] "IFNg_vs_naive" "salm_vs_naive" "ifng_vs_naive"It is possible to directly access specific DEA results either in
their original format or in minimal format. Use
format = "original" to retrieve results in the same class
as originally provided (e.g., DESeqResults,
MArrayLM…).
# access the 1st DEA if dea_name is not specified (default: minimal format)
dea(dde) |> head()
#> DataFrame with 6 rows and 3 columns
#> IFNg_vs_naive_log2FoldChange IFNg_vs_naive_pvalue
#> <numeric> <numeric>
#> ENSG00000164741 0.2291487 9.80541e-01
#> ENSG00000078808 -0.0115336 1.00000e+00
#> ENSG00000251034 -0.1167013 9.33899e-01
#> ENSG00000162676 0.2691481 9.00817e-01
#> ENSG00000170356 -0.1278637 9.72815e-01
#> ENSG00000204257 4.0550244 1.67298e-56
#> IFNg_vs_naive_padj
#> <numeric>
#> ENSG00000164741 1.00000e+00
#> ENSG00000078808 1.00000e+00
#> ENSG00000251034 1.00000e+00
#> ENSG00000162676 1.00000e+00
#> ENSG00000170356 1.00000e+00
#> ENSG00000204257 8.36489e-54
# access the 1st DEA, in original format
dea(dde, format = "original") |> head()
#> log2 fold change (MAP): condition IFNg vs naive
#> Wald test p-value: condition IFNg vs naive
#> DataFrame with 6 rows and 5 columns
#> baseMean log2FoldChange lfcSE pvalue padj
#> <numeric> <numeric> <numeric> <numeric> <numeric>
#> ENSG00000164741 918.0347 0.2291487 0.3006751 9.80541e-01 1.00000e+00
#> ENSG00000078808 6684.4755 -0.0115336 0.0729923 1.00000e+00 1.00000e+00
#> ENSG00000251034 12.1241 -0.1167013 0.4049828 9.33899e-01 1.00000e+00
#> ENSG00000162676 10.2220 0.2691481 0.3776971 9.00817e-01 1.00000e+00
#> ENSG00000170356 15.0772 -0.1278637 0.3522078 9.72815e-01 1.00000e+00
#> ENSG00000204257 1922.0256 4.0550244 0.1960055 1.67298e-56 8.36489e-54
# access specific DEA by name, in original format
dea(dde,
dea_name = "lrt_IFNg_vs_naive",
format = "original"
) |> head()
#> An object of class "DGELRT"
#> $coefficients
#> (Intercept) lineeiwy_1 linefikt_3 lineieki_2 linepodx_1
#> ENSG00000164741 5.175865 0.67213545 -0.05261294 -0.0133961 1.29681797
#> ENSG00000078808 8.278650 0.00402444 -0.03709639 0.2193207 0.11747661
#> ENSG00000251034 1.015714 0.14916999 0.74784074 0.2537142 0.06875325
#> ENSG00000162676 1.290001 0.33780292 0.40311584 -0.3190235 0.45653599
#> ENSG00000170356 2.727577 0.72922538 -3.77847802 -1.1287036 0.22635415
#> ENSG00000204257 5.014917 0.06181123 0.55205972 0.4028221 0.02654140
#> lineqaqx_1 groupIFNg groupIFNg_SL1344 groupSL1344
#> ENSG00000164741 2.6464968 0.209417086 0.2091795 0.008356587
#> ENSG00000078808 -0.1422928 -0.008349706 0.4510976 0.549873639
#> ENSG00000251034 -0.9196304 -0.151903783 2.0529807 0.936175420
#> ENSG00000162676 -0.1853207 0.281787362 1.3643530 0.367163846
#> ENSG00000170356 -1.2678875 -0.134828422 0.3154840 -0.490165844
#> ENSG00000204257 -0.2265600 2.824349649 2.8414754 -0.525496515
#>
#> $fitted.values
#> SAMEA103885102 SAMEA103885347 SAMEA103885043 SAMEA103885392
#> ENSG00000164741 331.659348 462.046537 263.439048 254.90142
#> ENSG00000078808 6870.927163 6678.847968 8959.634080 6786.72570
#> ENSG00000251034 4.911316 4.108220 9.209275 23.90284
#> ENSG00000162676 6.482512 8.484501 6.859816 15.69813
#> ENSG00000170356 31.760710 28.596699 17.917217 24.65952
#> ENSG00000204257 257.670764 4470.359901 116.241178 2839.36715
#> SAMEA103885182 SAMEA103885136 SAMEA103885413 SAMEA103884967
#> ENSG00000164741 541.666783 683.549644 512.978954 304.27088
#> ENSG00000078808 6344.398721 5486.053836 8924.936535 5199.85122
#> ENSG00000251034 5.156450 3.844418 10.586591 20.81102
#> ENSG00000162676 8.343854 9.757319 9.622103 16.69806
#> ENSG00000170356 59.995111 46.992545 29.035757 28.28534
#> ENSG00000204257 267.284775 3939.199536 119.973193 2260.61906
#> SAMEA103885368 SAMEA103885218 SAMEA103885319 SAMEA103885004
#> ENSG00000164741 359.1690921 296.1160031 185.9295535 135.7289205
#> ENSG00000078808 6581.0582108 4732.6215141 6470.3658204 4342.1495644
#> ENSG00000251034 10.6777230 6.5701908 15.2973496 33.3529553
#> ENSG00000162676 9.7702877 9.3993397 8.1610912 15.4157380
#> ENSG00000170356 0.4487317 0.2805971 0.1502375 0.1401802
#> ENSG00000204257 458.8311804 5723.0986920 152.9443903 3171.1674140
#> SAMEA103885284 SAMEA103885059 SAMEA103884898 SAMEA103885157
#> ENSG00000164741 198.853936 262.449336 157.717118 121.183076
#> ENSG00000078808 7091.287572 5612.862339 7289.093283 4792.280494
#> ENSG00000251034 4.657825 3.164586 7.226507 15.870678
#> ENSG00000162676 3.751739 3.996927 3.242135 6.341809
#> ENSG00000170356 3.740996 5.307562 3.444240 3.875783
#> ENSG00000204257 324.879377 4257.313691 110.558321 2388.251566
#> SAMEA103885111 SAMEA103884919 SAMEA103885276 SAMEA103885021
#> ENSG00000164741 1116.184441 804.017037 654.325856 584.52823
#> ENSG00000078808 7292.392451 5268.451796 9534.173331 5255.72692
#> ENSG00000251034 4.557560 2.918362 8.779133 15.94075
#> ENSG00000162676 4.360822 9.586404 10.152794 17.05224
#> ENSG00000170356 35.403402 24.459121 18.105985 18.48645
#> ENSG00000204257 251.787138 3039.052578 109.751067 1860.49582
#> SAMEA103885262 SAMEA103885228 SAMEA103885308 SAMEA103884949
#> ENSG00000164741 4676.035368 5469.611891 3263.933831 3151.881218
#> ENSG00000078808 7507.928126 6225.810693 8557.053697 5724.072261
#> ENSG00000251034 2.284445 1.626949 3.867357 9.080686
#> ENSG00000162676 6.722218 7.483029 6.310215 13.572598
#> ENSG00000170356 4.306305 7.589192 2.121924 7.343250
#> ENSG00000204257 276.880941 3841.057200 105.112020 2239.376276
#>
#> $deviance
#> [1] 17.904070 5.291987 16.516572 12.066070 19.281687 14.519999
#>
#> $iter
#> [1] 8 4 8 7 8 9
#>
#> $failed
#> [1] FALSE FALSE FALSE FALSE FALSE FALSE
#>
#> $method
#> [1] "levenberg"
#>
#> $unshrunk.coefficients
#> (Intercept) lineeiwy_1 linefikt_3 lineieki_2 linepodx_1
#> ENSG00000164741 5.386068 0.672387611 -0.05266479 -0.01339855 1.29719043
#> ENSG00000078808 8.492342 0.004024619 -0.03709664 0.21932448 0.11747876
#> ENSG00000251034 1.195414 0.148687976 0.75562359 0.25519469 0.06707072
#> ENSG00000162676 1.477191 0.341309626 0.40776075 -0.32582662 0.46188517
#> ENSG00000170356 2.928727 0.732201452 -4.12611348 -1.14264160 0.22784660
#> ENSG00000204257 5.226853 0.061876190 0.55234530 0.40296198 0.02654786
#> lineqaqx_1 groupIFNg groupIFNg_SL1344 groupSL1344
#> ENSG00000164741 2.6469780 0.20948778 0.2093696 0.008408254
#> ENSG00000078808 -0.1422954 -0.00834996 0.4511061 0.549883572
#> ENSG00000251034 -0.9495303 -0.15717553 2.0845273 0.956233102
#> ENSG00000162676 -0.1890058 0.28821392 1.3819415 0.374778403
#> ENSG00000170356 -1.2851835 -0.13693313 0.3174214 -0.496972239
#> ENSG00000204257 -0.2267053 2.82491005 2.8420494 -0.525893398
#>
#> $df.residual
#> [1] 15 15 15 15 15 15
#>
#> $design
#> (Intercept) lineeiwy_1 linefikt_3 lineieki_2 linepodx_1
#> SAMEA103885102 1 0 0 0 0
#> SAMEA103885347 1 0 0 0 0
#> SAMEA103885043 1 0 0 0 0
#> SAMEA103885392 1 0 0 0 0
#> SAMEA103885182 1 1 0 0 0
#> lineqaqx_1 groupIFNg groupIFNg_SL1344 groupSL1344
#> SAMEA103885102 0 0 0 0
#> SAMEA103885347 0 1 0 0
#> SAMEA103885043 0 0 0 1
#> SAMEA103885392 0 0 1 0
#> SAMEA103885182 0 0 0 0
#> 19 more rows ...
#>
#> $offset
#> [,1] [,2] [,3] [,4] [,5] [,6] [,7]
#> [1,] 0.4180404 0.5401098 0.17934576 -0.05456072 0.2361954 0.2593559 0.17337094
#> [2,] 0.3427124 0.3227088 0.05825912 -0.12072419 0.2589610 0.1219479 0.05035433
#> [3,] 0.3961281 0.3747517 0.06856426 -0.10594365 0.2961466 0.1596962 0.05925339
#> [4,] 0.3919171 0.3728362 0.07371127 -0.10559112 0.3030246 0.1713031 0.07078391
#> [5,] 0.5295032 0.5614976 0.45400752 -0.04098516 0.4333348 0.3259938 0.20457207
#> [6,] 0.3248296 0.3534611 0.05470751 -0.11756594 0.2995854 0.1650935 0.02443249
#> [,8] [,9] [,10] [,11] [,12] [,13]
#> [1,] -0.5499069 0.5503901 0.14786028 -0.1164436 -0.6321131 -0.08009890
#> [2,] -0.3910874 0.3367055 0.01533919 -0.2301410 -0.5302266 0.15495575
#> [3,] -0.3931466 0.4171223 0.08868107 -0.1795908 -0.5284183 0.08794008
#> [4,] -0.3851496 0.3943942 0.06747383 -0.1603522 -0.5315043 0.17085523
#> [5,] -0.6360060 0.3960564 0.06348445 -0.2011793 -1.0848610 -0.46673327
#> [6,] -0.4073846 0.3494840 0.04815727 -0.2232306 -0.5593926 0.15363894
#> [,14] [,15] [,16] [,17] [,18] [,19]
#> [1,] -0.01209918 -0.32027468 -0.7847366 0.3344130 -0.20312576 -0.208061219
#> [2,] -0.07050040 -0.36741559 -0.6880109 0.2847662 -0.03197896 0.002933508
#> [3,] -0.14141082 -0.42908574 -0.7706625 0.2543029 -0.03428650 -0.046339940
#> [4,] -0.05405233 -0.34991078 -0.6861418 -0.4664154 0.03305583 0.003894374
#> [5,] 0.01998052 -0.05240958 -0.7487589 0.4102345 0.17736293 0.236641356
#> [6,] -0.09833143 -0.39837846 -0.6935476 0.2751831 -0.05900987 -0.029292735
#> [,20] [,21] [,22] [,23] [,24]
#> [1,] -0.5218229 0.4171598 0.3644291 0.04923413 -0.18666088
#> [2,] -0.4938532 0.5736682 0.3947623 0.15458104 -0.14871694
#> [3,] -0.5781334 0.5802395 0.3979984 0.15045482 -0.12426103
#> [4,] -0.4847359 0.6172329 0.4362385 0.17920613 -0.06207378
#> [5,] -0.5569570 -0.1834631 0.5201149 -0.39424768 0.03281681
#> [6,] -0.5668520 0.6234398 0.4284451 0.18077229 -0.12824446
#> attr(,"class")
#> [1] "CompressedMatrix"
#> attr(,"Dims")
#> [1] 6 24
#> attr(,"repeat.row")
#> [1] FALSE
#> attr(,"repeat.col")
#> [1] FALSE
#>
#> $dispersion
#> [1] 0.15434109 0.01541182 0.14534478 0.11348545 0.11524396 0.05153858
#>
#> $prior.count
#> [1] 0.125
#>
#> $samples
#> group lib.size norm.factors names sample_id
#> SAMEA103885102 naive 2451140 1 SAMEA103885102 diku_A
#> SAMEA103885347 IFNg 2319742 1 SAMEA103885347 diku_B
#> SAMEA103885043 SL1344 2275254 1 SAMEA103885043 diku_C
#> SAMEA103885392 IFNg_SL1344 2041202 1 SAMEA103885392 diku_D
#> SAMEA103885182 naive 2415265 1 SAMEA103885182 eiwy_A
#> line_id replicate condition_name macrophage_harvest
#> SAMEA103885102 diku_1 1 naive 11/6/2015
#> SAMEA103885347 diku_1 1 IFNg 11/6/2015
#> SAMEA103885043 diku_1 1 SL1344 11/6/2015
#> SAMEA103885392 diku_1 1 IFNg_SL1344 11/6/2015
#> SAMEA103885182 eiwy_1 1 naive 11/25/2015
#> salmonella_date ng_ul_mean rna_extraction rna_submit
#> SAMEA103885102 11/13/2015 293.9625 11/30/2015 12/9/2015
#> SAMEA103885347 11/13/2015 293.9625 11/30/2015 12/9/2015
#> SAMEA103885043 11/13/2015 293.9625 11/30/2015 12/9/2015
#> SAMEA103885392 11/13/2015 293.9625 11/30/2015 12/9/2015
#> SAMEA103885182 12/2/2015 193.5450 12/3/2015 12/9/2015
#> library_pool chemistry rna_auto line
#> SAMEA103885102 3226_RNAauto-091215 V4_auto 1 diku_1
#> SAMEA103885347 3226_RNAauto-091215 V4_auto 1 diku_1
#> SAMEA103885043 3226_RNAauto-091215 V4_auto 1 diku_1
#> SAMEA103885392 3226_RNAauto-091215 V4_auto 1 diku_1
#> SAMEA103885182 3226_RNAauto-091215 V4_auto 1 eiwy_1
#> 19 more rows ...
#>
#> $genes
#> seqnames start end width strand gene_id
#> ENSG00000164741 chr8 13083361 13604610 521250 - ENSG00000164741.14
#> ENSG00000078808 chr1 1216908 1232031 15124 - ENSG00000078808.16
#> ENSG00000251034 chr8 22540845 22545405 4561 - ENSG00000251034.1
#> ENSG00000162676 chr1 92474762 92486876 12115 - ENSG00000162676.11
#> ENSG00000170356 chr7 144250045 144256244 6200 - ENSG00000170356.9
#> ENSG00000204257 chr6 32948613 32969094 20482 - ENSG00000204257.14
#> SYMBOL baseMean baseVar allZero dispGeneEst
#> ENSG00000164741 DLC1 918.03469 1.919478e+06 FALSE 0.176900497
#> ENSG00000078808 SDF4 6684.47548 4.121476e+06 FALSE 0.005336141
#> ENSG00000251034 <NA> 12.12408 2.083055e+02 FALSE 0.139939060
#> ENSG00000162676 GFI1 10.22204 8.849548e+01 FALSE 0.088009082
#> ENSG00000170356 OR2A20P 15.07716 2.865438e+02 FALSE 0.086413772
#> ENSG00000204257 HLA-DMA 1922.02562 3.556107e+06 FALSE 0.050271572
#> dispGeneIter dispFit dispersion dispIter dispOutlier
#> ENSG00000164741 8 0.07822483 0.16262236 10 FALSE
#> ENSG00000078808 8 0.07420142 0.00763372 9 FALSE
#> ENSG00000251034 13 0.42671547 0.19197295 8 FALSE
#> ENSG00000162676 30 0.49242762 0.14920745 6 FALSE
#> ENSG00000170356 8 0.35754498 0.15108319 8 FALSE
#> ENSG00000204257 8 0.07578857 0.05282007 7 FALSE
#> dispMAP Intercept line_eiwy_1_vs_diku_1
#> ENSG00000164741 0.16262236 7.770353 0.970022375
#> ENSG00000078808 0.00763372 12.251844 0.005781728
#> ENSG00000251034 0.19197295 1.755162 0.175340690
#> ENSG00000162676 0.14920745 2.136581 0.480446534
#> ENSG00000170356 0.15108319 4.233279 1.056913201
#> ENSG00000204257 0.05282007 7.540662 0.089423375
#> line_fikt_3_vs_diku_1 line_ieki_2_vs_diku_1
#> ENSG00000164741 -0.07577429 -0.01929912
#> ENSG00000078808 -0.05342186 0.31633010
#> ENSG00000251034 1.05452572 0.33475857
#> ENSG00000162676 0.58406093 -0.47081704
#> ENSG00000170356 -5.53704332 -1.66265291
#> ENSG00000204257 0.79727383 0.58138974
#> line_podx_1_vs_diku_1 line_qaqx_1_vs_diku_1
#> ENSG00000164741 1.87146580 3.8188294
#> ENSG00000078808 0.16953519 -0.2053341
#> ENSG00000251034 0.05036760 -1.4165928
#> ENSG00000162676 0.66518266 -0.2720575
#> ENSG00000170356 0.32751605 -1.8548902
#> ENSG00000204257 0.03820932 -0.3272907
#> condition_IFNg_vs_naive condition_IFNg_SL1344_vs_naive
#> ENSG00000164741 0.30218964 0.3025714
#> ENSG00000078808 -0.01205631 0.6508812
#> ENSG00000251034 -0.22261594 3.0195724
#> ENSG00000162676 0.42160612 1.9873533
#> ENSG00000170356 -0.19625391 0.4465885
#> ENSG00000204257 4.07555133 4.1002828
#> condition_SL1344_vs_naive SE_Intercept SE_line_eiwy_1_vs_diku_1
#> ENSG00000164741 0.0121682 0.35946727 0.41475882
#> ENSG00000078808 0.7933721 0.07791249 0.09000068
#> ENSG00000251034 1.3720821 0.52662721 0.57749189
#> ENSG00000162676 0.5395388 0.47091496 0.51421255
#> ENSG00000170356 -0.7301748 0.40909698 0.43816712
#> ENSG00000204257 -0.7587539 0.20930500 0.24109955
#> SE_line_fikt_3_vs_diku_1 SE_line_ieki_2_vs_diku_1
#> ENSG00000164741 0.41637071 0.41707883
#> ENSG00000078808 0.09007559 0.09001577
#> ENSG00000251034 0.55423062 0.59092538
#> ENSG00000162676 0.51540968 0.58148644
#> ENSG00000170356 1.08119676 0.55495255
#> ENSG00000204257 0.24021347 0.24106904
#> SE_line_podx_1_vs_diku_1 SE_line_qaqx_1_vs_diku_1
#> ENSG00000164741 0.41426348 0.41357906
#> ENSG00000078808 0.08998452 0.08995828
#> ENSG00000251034 0.58993435 0.64576993
#> ENSG00000162676 0.52198431 0.52861599
#> ENSG00000170356 0.44840747 0.53506242
#> ENSG00000204257 0.24138447 0.24133054
#> SE_condition_IFNg_vs_naive SE_condition_IFNg_SL1344_vs_naive
#> ENSG00000164741 0.33811564 0.33917339
#> ENSG00000078808 0.07354079 0.07356888
#> ENSG00000251034 0.55185235 0.47411418
#> ENSG00000162676 0.45132259 0.43087159
#> ENSG00000170356 0.42631745 0.43364895
#> ENSG00000204257 0.19471168 0.19486534
#> SE_condition_SL1344_vs_naive WaldStatistic_Intercept
#> ENSG00000164741 0.33870933 21.616302
#> ENSG00000078808 0.07340909 157.251345
#> ENSG00000251034 0.49674549 3.332836
#> ENSG00000162676 0.45764794 4.537084
#> ENSG00000170356 0.44708739 10.347861
#> ENSG00000204257 0.20193856 36.027147
#> WaldStatistic_line_eiwy_1_vs_diku_1
#> ENSG00000164741 2.33876251
#> ENSG00000078808 0.06424094
#> ENSG00000251034 0.30362451
#> ENSG00000162676 0.93433451
#> ENSG00000170356 2.41212351
#> ENSG00000204257 0.37089814
#> WaldStatistic_line_fikt_3_vs_diku_1
#> ENSG00000164741 -0.1819876
#> ENSG00000078808 -0.5930781
#> ENSG00000251034 1.9026840
#> ENSG00000162676 1.1331974
#> ENSG00000170356 -5.1212171
#> ENSG00000204257 3.3190221
#> WaldStatistic_line_ieki_2_vs_diku_1
#> ENSG00000164741 -0.04627211
#> ENSG00000078808 3.51416314
#> ENSG00000251034 0.56649888
#> ENSG00000162676 -0.80967846
#> ENSG00000170356 -2.99602717
#> ENSG00000204257 2.41171465
#> WaldStatistic_line_podx_1_vs_diku_1
#> ENSG00000164741 4.51757370
#> ENSG00000078808 1.88404850
#> ENSG00000251034 0.08537831
#> ENSG00000162676 1.27433458
#> ENSG00000170356 0.73039829
#> ENSG00000204257 0.15829236
#> WaldStatistic_line_qaqx_1_vs_diku_1
#> ENSG00000164741 9.233614
#> ENSG00000078808 -2.282548
#> ENSG00000251034 -2.193649
#> ENSG00000162676 -0.514660
#> ENSG00000170356 -3.466680
#> ENSG00000204257 -1.356193
#> WaldStatistic_condition_IFNg_vs_naive
#> ENSG00000164741 0.8937464
#> ENSG00000078808 -0.1639404
#> ENSG00000251034 -0.4033977
#> ENSG00000162676 0.9341569
#> ENSG00000170356 -0.4603469
#> ENSG00000204257 20.9312117
#> WaldStatistic_condition_IFNg_SL1344_vs_naive
#> ENSG00000164741 0.8920846
#> ENSG00000078808 8.8472347
#> ENSG00000251034 6.3688717
#> ENSG00000162676 4.6124028
#> ENSG00000170356 1.0298387
#> ENSG00000204257 21.0416218
#> WaldStatistic_condition_SL1344_vs_naive WaldPvalue_Intercept
#> ENSG00000164741 0.03592521 1.261922e-103
#> ENSG00000078808 10.80754512 0.000000e+00
#> ENSG00000251034 2.76214305 8.596573e-04
#> ENSG00000162676 1.17893863 5.703737e-06
#> ENSG00000170356 -1.63318139 4.279378e-25
#> ENSG00000204257 -3.75735038 3.144547e-284
#> WaldPvalue_line_eiwy_1_vs_diku_1
#> ENSG00000164741 0.01934773
#> ENSG00000078808 0.94877838
#> ENSG00000251034 0.76141398
#> ENSG00000162676 0.35013137
#> ENSG00000170356 0.01585991
#> ENSG00000204257 0.71071340
#> WaldPvalue_line_fikt_3_vs_diku_1
#> ENSG00000164741 8.555925e-01
#> ENSG00000078808 5.531289e-01
#> ENSG00000251034 5.708179e-02
#> ENSG00000162676 2.571313e-01
#> ENSG00000170356 3.035699e-07
#> ENSG00000204257 9.033327e-04
#> WaldPvalue_line_ieki_2_vs_diku_1
#> ENSG00000164741 0.9630933700
#> ENSG00000078808 0.0004411418
#> ENSG00000251034 0.5710546971
#> ENSG00000162676 0.4181250005
#> ENSG00000170356 0.0027352206
#> ENSG00000204257 0.0158777027
#> WaldPvalue_line_podx_1_vs_diku_1
#> ENSG00000164741 6.255226e-06
#> ENSG00000078808 5.955842e-02
#> ENSG00000251034 9.319606e-01
#> ENSG00000162676 2.025449e-01
#> ENSG00000170356 4.651468e-01
#> ENSG00000204257 8.742264e-01
#> WaldPvalue_line_qaqx_1_vs_diku_1
#> ENSG00000164741 2.616515e-20
#> ENSG00000078808 2.245698e-02
#> ENSG00000251034 2.826063e-02
#> ENSG00000162676 6.067907e-01
#> ENSG00000170356 5.269289e-04
#> ENSG00000204257 1.750379e-01
#> WaldPvalue_condition_IFNg_vs_naive
#> ENSG00000164741 3.714576e-01
#> ENSG00000078808 8.697780e-01
#> ENSG00000251034 6.866557e-01
#> ENSG00000162676 3.502229e-01
#> ENSG00000170356 6.452673e-01
#> ENSG00000204257 2.783309e-97
#> WaldPvalue_condition_IFNg_SL1344_vs_naive
#> ENSG00000164741 3.723476e-01
#> ENSG00000078808 8.971461e-19
#> ENSG00000251034 1.904239e-10
#> ENSG00000162676 3.980406e-06
#> ENSG00000170356 3.030857e-01
#> ENSG00000204257 2.728839e-98
#> WaldPvalue_condition_SL1344_vs_naive betaConv betaIter deviance
#> ENSG00000164741 9.713420e-01 TRUE 5 315.9203
#> ENSG00000078808 3.170408e-27 TRUE 3 358.9627
#> ENSG00000251034 5.742331e-03 TRUE 5 124.7960
#> ENSG00000162676 2.384226e-01 TRUE 5 124.1491
#> ENSG00000170356 1.024309e-01 TRUE 9 129.6075
#> ENSG00000204257 1.717220e-04 TRUE 4 307.6203
#> maxCooks
#> ENSG00000164741 NA
#> ENSG00000078808 NA
#> ENSG00000251034 NA
#> ENSG00000162676 NA
#> ENSG00000170356 NA
#> ENSG00000204257 NA
#>
#> $prior.df
#> [1] 3.944878
#>
#> $AveLogCPM
#> [1] 9.002025 11.648884 2.516565 2.454987 3.149487 9.783568
#>
#> $table
#> logFC logCPM LR PValue
#> ENSG00000164741 0.30212499 9.002025 0.83286135 3.614464e-01
#> ENSG00000078808 -0.01204608 11.648884 0.01342048 9.077740e-01
#> ENSG00000251034 -0.21915083 2.516565 0.18895469 6.637880e-01
#> ENSG00000162676 0.40653323 2.454987 0.95277969 3.290128e-01
#> ENSG00000170356 -0.19451630 3.149487 0.24825672 6.183053e-01
#> ENSG00000204257 4.07467523 9.783568 353.92885191 5.910033e-79
#>
#> $comparison
#> [1] "1*groupIFNg"
#>
#> $df.test
#> [1] 1 1 1 1 1 1
# access specific DEA by name (default: minimal format)
dea(dde, dea_name = "lrt_IFNg_vs_naive") |> head()
#> DataFrame with 6 rows and 3 columns
#> lrt_IFNg_vs_naive_log2FoldChange lrt_IFNg_vs_naive_pvalue
#> <numeric> <numeric>
#> ENSG00000164741 0.3021250 3.61446e-01
#> ENSG00000078808 -0.0120461 9.07774e-01
#> ENSG00000251034 -0.2191508 6.63788e-01
#> ENSG00000162676 0.4065332 3.29013e-01
#> ENSG00000170356 -0.1945163 6.18305e-01
#> ENSG00000204257 4.0746752 5.91003e-79
#> lrt_IFNg_vs_naive_padj
#> <numeric>
#> ENSG00000164741 5.49722e-01
#> ENSG00000078808 9.53613e-01
#> ENSG00000251034 8.00709e-01
#> ENSG00000162676 5.19240e-01
#> ENSG00000170356 7.76765e-01
#> ENSG00000204257 5.91003e-76get_dea_list() retrieves all available DEAs stored in a
dde object as a list.
# get dea results as a list, (default: minimal format)
lapply(get_dea_list(dde), head)
#> $IFNg_vs_naive
#> log2FoldChange pvalue padj
#> ENSG00000164741 0.22914867 9.805414e-01 1.000000e+00
#> ENSG00000078808 -0.01153363 1.000000e+00 1.000000e+00
#> ENSG00000251034 -0.11670132 9.338990e-01 1.000000e+00
#> ENSG00000162676 0.26914813 9.008170e-01 1.000000e+00
#> ENSG00000170356 -0.12786371 9.728148e-01 1.000000e+00
#> ENSG00000204257 4.05502442 1.672977e-56 8.364885e-54
#>
#> $Salm_vs_naive
#> log2FoldChange pvalue padj
#> ENSG00000164741 0.01146911 0.9996325 1.0000000
#> ENSG00000078808 0.78936008 0.9975592 1.0000000
#> ENSG00000251034 1.18814969 0.2269175 0.9820422
#> ENSG00000162676 0.44208214 0.8432117 1.0000000
#> ENSG00000170356 -0.62492007 0.7269723 1.0000000
#> ENSG00000204257 -0.72938097 0.8838883 1.0000000
#>
#> $lrt_IFNg_vs_naive
#> log2FoldChange pvalue padj
#> ENSG00000164741 0.30212499 3.614464e-01 5.497216e-01
#> ENSG00000078808 -0.01204608 9.077740e-01 9.536130e-01
#> ENSG00000251034 -0.21915083 6.637880e-01 8.007093e-01
#> ENSG00000162676 0.40653323 3.290128e-01 5.192398e-01
#> ENSG00000170356 -0.19451630 6.183053e-01 7.767654e-01
#> ENSG00000204257 4.07467523 5.910033e-79 5.910033e-76
#>
#> $same_contrast
#> log2FoldChange pvalue padj
#> ENSG00000164741 -0.7170764 4.140413e-01 5.742598e-01
#> ENSG00000078808 0.8006668 5.343686e-06 4.679862e-05
#> ENSG00000251034 1.3949969 3.018601e-02 7.840521e-02
#> ENSG00000162676 0.7150586 1.294292e-01 2.462148e-01
#> ENSG00000170356 -0.3280573 7.530547e-01 8.675746e-01
#> ENSG00000204257 -0.7804105 4.505584e-02 1.083073e-01The next command can be quite verbose in its output, but it gives
full access to the original objects provided when creating and updating
the dde container.
# get dea results as a list, in original format
lapply(get_dea_list(dde, format = "original"), head)dea_info() returns the whole content of the
dea slot, including the DEA tables and their associated
metadata - the following chunk is left unevaluated for clarity as its
output can be very verbose.
# extra info
dea_info(dde)It is easy, still, to retrieve some specific set of information, such
as the package used for a specific
dea_name:
# retrieve specific information like the package name used for specific dea
dea_name <- "Salm_vs_naive"
dea_info(dde)[[dea_name]][["package"]]
#> [1] "DESeq2"fea() directly accesses specific FEA results either in
their original format or in minimal format
# access the 1st FEA, (default: minimal format)
fea(dde) |> head()
#> gs_id
#> GO:0002822 GO:0002822
#> GO:0002697 GO:0002697
#> GO:0002250 GO:0002250
#> GO:1904064 GO:1904064
#> GO:0042098 GO:0042098
#> GO:0070663 GO:0070663
#> gs_description
#> GO:0002822 regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
#> GO:0002697 regulation of immune effector process
#> GO:0002250 adaptive immune response
#> GO:1904064 positive regulation of cation transmembrane transport
#> GO:0042098 T cell proliferation
#> GO:0070663 regulation of leukocyte proliferation
#> gs_pvalue
#> GO:0002822 3.90e-06
#> GO:0002697 1.30e-05
#> GO:0002250 3.70e-05
#> GO:1904064 8.80e-04
#> GO:0042098 8.80e-04
#> GO:0070663 1.34e-03
#> gs_genes
#> GO:0002822 CD28,CD69,CR1L,IL27,SECTM1,TNFSF18
#> GO:0002697 CD28,CD69,CR1L,IL27,SECTM1,TNFSF18
#> GO:0002250 CD28,CD69,CR1L,CTSS,HLA-DMA,HLA-DPA1,IL27,LAMP3,SECTM1,TNFSF18
#> GO:1904064 ANK2,CTSS,KCNE5,P2RX4
#> GO:0042098 CD28,HLA-DPA1,IL27,TNFSF18
#> GO:0070663 CD28,HLA-DPA1,IL27,TNFSF18
#> gs_de_count gs_bg_count Expected
#> GO:0002822 6 11 0.55
#> GO:0002697 6 13 0.65
#> GO:0002250 10 17 0.85
#> GO:1904064 4 10 0.50
#> GO:0042098 4 10 0.50
#> GO:0070663 4 11 0.55
# access the 1st FEA, in original format
fea(dde, format = "original") |> head()
#> GO.ID
#> 1 GO:0002822
#> 2 GO:0002697
#> 3 GO:0002250
#> 4 GO:1904064
#> 5 GO:0042098
#> 6 GO:0070663
#> Term
#> 1 regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
#> 2 regulation of immune effector process
#> 3 adaptive immune response
#> 4 positive regulation of cation transmembrane transport
#> 5 T cell proliferation
#> 6 regulation of leukocyte proliferation
#> Annotated Significant Expected Rank in p.value_classic p.value_elim
#> 1 11 6 0.55 2 3.90e-06
#> 2 13 6 0.65 5 1.30e-05
#> 3 17 10 0.85 1 3.70e-05
#> 4 10 4 0.50 13 8.80e-04
#> 5 10 4 0.50 14 8.80e-04
#> 6 11 4 0.55 17 1.34e-03
#> p.value_classic
#> 1 3.90e-06
#> 2 1.30e-05
#> 3 4.00e-10
#> 4 8.80e-04
#> 5 8.80e-04
#> 6 1.34e-03
#> genes
#> 1 CD28,CD69,CR1L,IL27,SECTM1,TNFSF18
#> 2 CD28,CD69,CR1L,IL27,SECTM1,TNFSF18
#> 3 CD28,CD69,CR1L,CTSS,HLA-DMA,HLA-DPA1,IL27,LAMP3,SECTM1,TNFSF18
#> 4 ANK2,CTSS,KCNE5,P2RX4
#> 5 CD28,HLA-DPA1,IL27,TNFSF18
#> 6 CD28,HLA-DPA1,IL27,TNFSF18
# access specific FEA by name
fea(dde, fea_name = "ifng_vs_naive") |> head()
#> gs_id
#> GO:0002250 GO:0002250
#> GO:0002819 GO:0002819
#> GO:0002822 GO:0002822
#> GO:0002460 GO:0002460
#> GO:0002697 GO:0002697
#> GO:0050776 GO:0050776
#> gs_description
#> GO:0002250 adaptive immune response
#> GO:0002819 regulation of adaptive immune response
#> GO:0002822 regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
#> GO:0002460 adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
#> GO:0002697 regulation of immune effector process
#> GO:0050776 regulation of immune response
#> gs_pvalue
#> GO:0002250 3.958927e-10
#> GO:0002819 3.872646e-06
#> GO:0002822 3.872646e-06
#> GO:0002460 7.465887e-06
#> GO:0002697 1.336486e-05
#> GO:0050776 7.666847e-05
#> gs_genes
#> GO:0002250 ENSG00000231389,ENSG00000204257,ENSG00000078081,ENSG00000141574,ENSG00000197721,ENSG00000178562,ENSG00000197272,ENSG00000120337,ENSG00000163131,ENSG00000110848
#> GO:0002819 ENSG00000141574,ENSG00000197721,ENSG00000178562,ENSG00000197272,ENSG00000120337,ENSG00000110848
#> GO:0002822 ENSG00000141574,ENSG00000197721,ENSG00000178562,ENSG00000197272,ENSG00000120337,ENSG00000110848
#> GO:0002460 ENSG00000141574,ENSG00000197721,ENSG00000178562,ENSG00000197272,ENSG00000120337,ENSG00000110848
#> GO:0002697 ENSG00000141574,ENSG00000197721,ENSG00000178562,ENSG00000197272,ENSG00000120337,ENSG00000110848
#> GO:0050776 ENSG00000231389,ENSG00000204257,ENSG00000141574,ENSG00000197721,ENSG00000178562,ENSG00000197272,ENSG00000120337,ENSG00000163131,ENSG00000110848
#> gs_de_count gs_bg_count gs_ontology GeneRatio BgRatio p.adjust
#> GO:0002250 10 17 BP 10/37 17/744 2.561426e-07
#> GO:0002819 6 11 BP 6/37 11/744 8.352007e-04
#> GO:0002822 6 11 BP 6/37 11/744 8.352007e-04
#> GO:0002460 6 12 BP 6/37 12/744 1.207607e-03
#> GO:0002697 6 13 BP 6/37 13/744 1.729413e-03
#> GO:0050776 9 41 BP 9/37 41/744 8.267416e-03
#> qvalue
#> GO:0002250 2.233668e-07
#> GO:0002819 7.283293e-04
#> GO:0002822 7.283293e-04
#> GO:0002460 1.053083e-03
#> GO:0002697 1.508119e-03
#> GO:0050776 7.209526e-03
# access specific FEA by name, in original format
fea(dde, fea_name = "ifng_vs_naive", format = "original") |> head()
#> ID
#> GO:0002250 GO:0002250
#> GO:0002819 GO:0002819
#> GO:0002822 GO:0002822
#> GO:0002460 GO:0002460
#> GO:0002697 GO:0002697
#> GO:0050776 GO:0050776
#> Description
#> GO:0002250 adaptive immune response
#> GO:0002819 regulation of adaptive immune response
#> GO:0002822 regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
#> GO:0002460 adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
#> GO:0002697 regulation of immune effector process
#> GO:0050776 regulation of immune response
#> GeneRatio BgRatio RichFactor FoldEnrichment zScore pvalue
#> GO:0002250 10/37 17/744 0.5882353 11.828299 10.325309 3.958927e-10
#> GO:0002819 6/37 11/744 0.5454545 10.968059 7.614485 3.872646e-06
#> GO:0002822 6/37 11/744 0.5454545 10.968059 7.614485 3.872646e-06
#> GO:0002460 6/37 12/744 0.5000000 10.054054 7.228759 7.465887e-06
#> GO:0002697 6/37 13/744 0.4615385 9.280665 6.885949 1.336486e-05
#> GO:0050776 9/37 41/744 0.2195122 4.413975 5.141149 7.666847e-05
#> p.adjust qvalue
#> GO:0002250 2.561426e-07 2.233668e-07
#> GO:0002819 8.352007e-04 7.283293e-04
#> GO:0002822 8.352007e-04 7.283293e-04
#> GO:0002460 1.207607e-03 1.053083e-03
#> GO:0002697 1.729413e-03 1.508119e-03
#> GO:0050776 8.267416e-03 7.209526e-03
#> geneID
#> GO:0002250 ENSG00000231389/ENSG00000204257/ENSG00000078081/ENSG00000141574/ENSG00000197721/ENSG00000178562/ENSG00000197272/ENSG00000120337/ENSG00000163131/ENSG00000110848
#> GO:0002819 ENSG00000141574/ENSG00000197721/ENSG00000178562/ENSG00000197272/ENSG00000120337/ENSG00000110848
#> GO:0002822 ENSG00000141574/ENSG00000197721/ENSG00000178562/ENSG00000197272/ENSG00000120337/ENSG00000110848
#> GO:0002460 ENSG00000141574/ENSG00000197721/ENSG00000178562/ENSG00000197272/ENSG00000120337/ENSG00000110848
#> GO:0002697 ENSG00000141574/ENSG00000197721/ENSG00000178562/ENSG00000197272/ENSG00000120337/ENSG00000110848
#> GO:0050776 ENSG00000231389/ENSG00000204257/ENSG00000141574/ENSG00000197721/ENSG00000178562/ENSG00000197272/ENSG00000120337/ENSG00000163131/ENSG00000110848
#> Count
#> GO:0002250 10
#> GO:0002819 6
#> GO:0002822 6
#> GO:0002460 6
#> GO:0002697 6
#> GO:0050776 9Similarly to get_dea_list(), get_fea_list()
returns all FEAs stored in a dde object. Optionally, if
dea_name is set, it returns all FEAs linked to a specific
DEA. This is illustrated in the following chunk
# get fea results as a list (default: minimal format)
lapply(get_fea_list(dde), head)
#> $IFNg_vs_naive
#> gs_id
#> GO:0002822 GO:0002822
#> GO:0002697 GO:0002697
#> GO:0002250 GO:0002250
#> GO:1904064 GO:1904064
#> GO:0042098 GO:0042098
#> GO:0070663 GO:0070663
#> gs_description
#> GO:0002822 regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
#> GO:0002697 regulation of immune effector process
#> GO:0002250 adaptive immune response
#> GO:1904064 positive regulation of cation transmembrane transport
#> GO:0042098 T cell proliferation
#> GO:0070663 regulation of leukocyte proliferation
#> gs_pvalue
#> GO:0002822 3.90e-06
#> GO:0002697 1.30e-05
#> GO:0002250 3.70e-05
#> GO:1904064 8.80e-04
#> GO:0042098 8.80e-04
#> GO:0070663 1.34e-03
#> gs_genes
#> GO:0002822 CD28,CD69,CR1L,IL27,SECTM1,TNFSF18
#> GO:0002697 CD28,CD69,CR1L,IL27,SECTM1,TNFSF18
#> GO:0002250 CD28,CD69,CR1L,CTSS,HLA-DMA,HLA-DPA1,IL27,LAMP3,SECTM1,TNFSF18
#> GO:1904064 ANK2,CTSS,KCNE5,P2RX4
#> GO:0042098 CD28,HLA-DPA1,IL27,TNFSF18
#> GO:0070663 CD28,HLA-DPA1,IL27,TNFSF18
#> gs_de_count gs_bg_count Expected
#> GO:0002822 6 11 0.55
#> GO:0002697 6 13 0.65
#> GO:0002250 10 17 0.85
#> GO:1904064 4 10 0.50
#> GO:0042098 4 10 0.50
#> GO:0070663 4 11 0.55
#>
#> $salm_vs_naive
#> gs_id gs_description
#> GO:0042221 GO:0042221 response to chemical
#> GO:0034097 GO:0034097 response to cytokine
#> GO:1901652 GO:1901652 response to peptide
#> GO:0070887 GO:0070887 cellular response to chemical stimulus
#> GO:0071356 GO:0071356 cellular response to tumor necrosis factor
#> GO:1901701 GO:1901701 cellular response to oxygen-containing compound
#> gs_pvalue
#> GO:0042221 0.01425000
#> GO:0034097 0.03856196
#> GO:1901652 0.03856196
#> GO:0070887 0.03904673
#> GO:0071356 0.39973004
#> GO:1901701 0.41892532
#> gs_genes
#> GO:0042221 LAMP3,RAPGEF2,IL12RB2,P2RX4,SHPK,CYP3A5,ADPRS,AGRN,TRERF1,SRXN1,UGCG,TNFSF18,RELA,NAIP,OSBPL7,CH25H,DNAJA1,MT1A,HAS2,PDE2A,SLC11A2,HAMP,KHK,GREM1,CCL2,LCOR,TSHR,GPR37,ADAR,OCSTAMP
#> GO:0034097 LAMP3,IL12RB2,SHPK,UGCG,TNFSF18,RELA,NAIP,CH25H,HAS2,PDE2A,CCL2,ADAR,OCSTAMP
#> GO:1901652 LAMP3,IL12RB2,SHPK,UGCG,TNFSF18,RELA,NAIP,CH25H,HAS2,PDE2A,CCL2,ADAR,OCSTAMP
#> GO:0070887 RAPGEF2,P2RX4,SHPK,CYP3A5,ADPRS,AGRN,TRERF1,SRXN1,UGCG,RELA,OSBPL7,CH25H,DNAJA1,MT1A,PDE2A,SLC11A2,GREM1,CCL2,LCOR,TSHR,GPR37,OCSTAMP
#> GO:0071356 TNFSF18,RELA,NAIP,HAS2,CCL2,OCSTAMP
#> GO:1901701 RAPGEF2,P2RX4,SHPK,ADPRS,AGRN,TRERF1,RELA,OSBPL7,PDE2A,CCL2,LCOR,TSHR,GPR37
#> gs_de_count gs_bg_count gs_adj_pvalue gs_ontology
#> GO:0042221 30 2551 0.01425000 GO:BP
#> GO:0034097 13 625 0.03856196 GO:BP
#> GO:1901652 13 625 0.03856196 GO:BP
#> GO:0070887 22 1616 0.03904673 GO:BP
#> GO:0071356 6 164 0.39973004 GO:BP
#> GO:1901701 13 791 0.41892532 GO:BP
#>
#> $ifng_vs_naive
#> gs_id
#> GO:0002250 GO:0002250
#> GO:0002819 GO:0002819
#> GO:0002822 GO:0002822
#> GO:0002460 GO:0002460
#> GO:0002697 GO:0002697
#> GO:0050776 GO:0050776
#> gs_description
#> GO:0002250 adaptive immune response
#> GO:0002819 regulation of adaptive immune response
#> GO:0002822 regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
#> GO:0002460 adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
#> GO:0002697 regulation of immune effector process
#> GO:0050776 regulation of immune response
#> gs_pvalue
#> GO:0002250 3.958927e-10
#> GO:0002819 3.872646e-06
#> GO:0002822 3.872646e-06
#> GO:0002460 7.465887e-06
#> GO:0002697 1.336486e-05
#> GO:0050776 7.666847e-05
#> gs_genes
#> GO:0002250 ENSG00000231389,ENSG00000204257,ENSG00000078081,ENSG00000141574,ENSG00000197721,ENSG00000178562,ENSG00000197272,ENSG00000120337,ENSG00000163131,ENSG00000110848
#> GO:0002819 ENSG00000141574,ENSG00000197721,ENSG00000178562,ENSG00000197272,ENSG00000120337,ENSG00000110848
#> GO:0002822 ENSG00000141574,ENSG00000197721,ENSG00000178562,ENSG00000197272,ENSG00000120337,ENSG00000110848
#> GO:0002460 ENSG00000141574,ENSG00000197721,ENSG00000178562,ENSG00000197272,ENSG00000120337,ENSG00000110848
#> GO:0002697 ENSG00000141574,ENSG00000197721,ENSG00000178562,ENSG00000197272,ENSG00000120337,ENSG00000110848
#> GO:0050776 ENSG00000231389,ENSG00000204257,ENSG00000141574,ENSG00000197721,ENSG00000178562,ENSG00000197272,ENSG00000120337,ENSG00000163131,ENSG00000110848
#> gs_de_count gs_bg_count gs_ontology GeneRatio BgRatio p.adjust
#> GO:0002250 10 17 BP 10/37 17/744 2.561426e-07
#> GO:0002819 6 11 BP 6/37 11/744 8.352007e-04
#> GO:0002822 6 11 BP 6/37 11/744 8.352007e-04
#> GO:0002460 6 12 BP 6/37 12/744 1.207607e-03
#> GO:0002697 6 13 BP 6/37 13/744 1.729413e-03
#> GO:0050776 9 41 BP 9/37 41/744 8.267416e-03
#> qvalue
#> GO:0002250 2.233668e-07
#> GO:0002819 7.283293e-04
#> GO:0002822 7.283293e-04
#> GO:0002460 1.053083e-03
#> GO:0002697 1.508119e-03
#> GO:0050776 7.209526e-03
# get all FEAs for this de_name, in original format
lapply(get_fea_list(dde, dea_name = "IFNg_vs_naive", format = "original"), head)
#> $IFNg_vs_naive
#> GO.ID
#> 1 GO:0002822
#> 2 GO:0002697
#> 3 GO:0002250
#> 4 GO:1904064
#> 5 GO:0042098
#> 6 GO:0070663
#> Term
#> 1 regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
#> 2 regulation of immune effector process
#> 3 adaptive immune response
#> 4 positive regulation of cation transmembrane transport
#> 5 T cell proliferation
#> 6 regulation of leukocyte proliferation
#> Annotated Significant Expected Rank in p.value_classic p.value_elim
#> 1 11 6 0.55 2 3.90e-06
#> 2 13 6 0.65 5 1.30e-05
#> 3 17 10 0.85 1 3.70e-05
#> 4 10 4 0.50 13 8.80e-04
#> 5 10 4 0.50 14 8.80e-04
#> 6 11 4 0.55 17 1.34e-03
#> p.value_classic
#> 1 3.90e-06
#> 2 1.30e-05
#> 3 4.00e-10
#> 4 8.80e-04
#> 5 8.80e-04
#> 6 1.34e-03
#> genes
#> 1 CD28,CD69,CR1L,IL27,SECTM1,TNFSF18
#> 2 CD28,CD69,CR1L,IL27,SECTM1,TNFSF18
#> 3 CD28,CD69,CR1L,CTSS,HLA-DMA,HLA-DPA1,IL27,LAMP3,SECTM1,TNFSF18
#> 4 ANK2,CTSS,KCNE5,P2RX4
#> 5 CD28,HLA-DPA1,IL27,TNFSF18
#> 6 CD28,HLA-DPA1,IL27,TNFSF18fea_info() returns the whole content of the
fea slot, including the FEA tables and their associated
metadata. Again, the following chunk is left unevaluated for clarity as
its output can be very verbose:
# extra info
fea_info(dde)Yet, specific set of information, such as the fe_tool
used for a specific fea_name (here, “ifng_vs_naive”) can be
easily retrieved with:
# the tool used to perform the FEA
fea_info(dde)[["ifng_vs_naive"]][["fe_tool"]]
#> [1] "clusterProfiler"Downstream operations with DeeDeeExperiment
objects
Since DeeDeeExperiment inherits from the
SummarizedExperiment class, it can be seamlessly used with
other Bioconductor tools such as iSEE or
GeneTonic, or many more.
With the dde object now containing all key elements of
your analysis, from the counts to enrichment results, metadata, and
scenario details, you can save it and plug it into these tools for
interactive exploration and visualization.
saveRDS(dde, "dde_macrophage.RDS")To explore the dde object within iSEE,
simply run these lines of code:
Similarly, to see more on dde within
GeneTonic, you can run the following chunk:
library("GeneTonic")
my_dde <- dde
res_de <- dea(my_dde, dea_name = "IFNg_vs_naive", format = "original")
res_enrich <- fea(my_dde, fea_name = "ifng_vs_naive")
annotation_object <- mosdef::get_annotation_orgdb(dds_macrophage,
"org.Hs.eg.db",
"ENSEMBL")
GeneTonic::GeneTonic(
dds = dds_macrophage,
res_de = res_de,
res_enrich = res_enrich,
annotation_obj = annotation_object,
project_id = "GeneTonic_with_DeeDee"
)Of course, these packages need to be installed as usual on the same
machine via BiocManager::install().
FAQs
Do I need to process/filter my DE results before adding them to a
dde object?
→ DeeDeeExperiment is just a container, it does not
alter your results. Any filtering or DEG selection should be done
beforehand, giving you full control over how your analysis is
conducted.
What happens if I forget to name my DE or FE result when adding it?
→ If no name is provided, DeeDeeExperiment will attempt
to assign a default one, which is the variable name in the current
environment. However, it’s strongly recommended to name each result
clearly.
I am using a different enrichment analysis tool, and this is currently not supported by DeeDeeExperiment, what can I do?
→ The quick-and-easy option would be to rename the output of the
tabular format from this tool into the format expected by
DeeDeeExperiment (which is the same adopted by
GeneTonic). If you think a conversion function would be
useful to yourself and many other users, feel free to submit a pull
request within the DeeDeeExperiment Github repository (https://github.com/imbeimainz/DeeDeeExperiment/pulls)!
Session info
sessionInfo()
#> R version 4.5.0 (2025-04-11)
#> Platform: aarch64-apple-darwin20
#> Running under: macOS Ventura 13.6
#>
#> Matrix products: default
#> BLAS: /Library/Frameworks/R.framework/Versions/4.5-arm64/Resources/lib/libRblas.0.dylib
#> LAPACK: /Library/Frameworks/R.framework/Versions/4.5-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.1
#>
#> locale:
#> [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
#>
#> time zone: Europe/Berlin
#> tzcode source: internal
#>
#> attached base packages:
#> [1] stats4 stats graphics grDevices utils datasets methods
#> [8] base
#>
#> other attached packages:
#> [1] DEFormats_1.36.0 edgeR_4.6.2
#> [3] limma_3.63.13 DESeq2_1.47.5
#> [5] macrophage_1.23.0 DeeDeeExperiment_0.99.0
#> [7] SummarizedExperiment_1.37.0 Biobase_2.67.0
#> [9] GenomicRanges_1.59.1 GenomeInfoDb_1.43.4
#> [11] IRanges_2.41.3 S4Vectors_0.45.4
#> [13] BiocGenerics_0.53.6 generics_0.1.3
#> [15] MatrixGenerics_1.19.1 matrixStats_1.5.0
#> [17] BiocStyle_2.35.0
#>
#> loaded via a namespace (and not attached):
#> [1] DBI_1.2.3 rlang_1.1.6 magrittr_2.0.3
#> [4] DOSE_4.1.0 compiler_4.5.0 RSQLite_2.3.9
#> [7] reshape2_1.4.4 png_0.1-8 systemfonts_1.2.2
#> [10] vctrs_0.6.5 stringr_1.5.1 pkgconfig_2.0.3
#> [13] crayon_1.5.3 fastmap_1.2.0 backports_1.5.0
#> [16] XVector_0.47.2 rmarkdown_2.29 UCSC.utils_1.3.1
#> [19] ragg_1.4.0 bit_4.6.0 xfun_0.52
#> [22] cachem_1.1.0 jsonlite_2.0.0 blob_1.2.4
#> [25] DelayedArray_0.33.6 BiocParallel_1.41.5 parallel_4.5.0
#> [28] R6_2.6.1 stringi_1.8.7 bslib_0.9.0
#> [31] jquerylib_0.1.4 GOSemSim_2.33.0 numDeriv_2016.8-1.1
#> [34] Rcpp_1.0.14 bookdown_0.42 knitr_1.50
#> [37] R.utils_2.13.0 Matrix_1.7-3 splines_4.5.0
#> [40] tidyselect_1.2.1 qvalue_2.39.0 rstudioapi_0.17.1
#> [43] abind_1.4-8 yaml_2.3.10 codetools_0.2-20
#> [46] lattice_0.22-7 tibble_3.2.1 plyr_1.8.9
#> [49] KEGGREST_1.47.1 coda_0.19-4.1 evaluate_1.0.3
#> [52] desc_1.4.3 Biostrings_2.75.4 pillar_1.10.2
#> [55] BiocManager_1.30.25 checkmate_2.3.2 emdbook_1.3.13
#> [58] ggplot2_3.5.2 munsell_0.5.1 scales_1.3.0
#> [61] glue_1.8.0 tools_4.5.0 apeglm_1.29.0
#> [64] data.table_1.17.0 fgsea_1.33.4 locfit_1.5-9.12
#> [67] fs_1.6.6 mvtnorm_1.3-3 fastmatch_1.1-6
#> [70] cowplot_1.1.3 grid_4.5.0 bbmle_1.0.25.1
#> [73] bdsmatrix_1.3-7 AnnotationDbi_1.69.1 colorspace_2.1-1
#> [76] GenomeInfoDbData_1.2.14 cli_3.6.5 textshaping_1.0.0
#> [79] S4Arrays_1.7.3 dplyr_1.1.4 gtable_0.3.6
#> [82] yulab.utils_0.2.0 R.methodsS3_1.8.2 sass_0.4.10
#> [85] digest_0.6.37 SparseArray_1.7.7 htmlwidgets_1.6.4
#> [88] R.oo_1.27.0 memoise_2.0.1 htmltools_0.5.8.1
#> [91] pkgdown_2.1.1 lifecycle_1.0.4 httr_1.4.7
#> [94] GO.db_3.21.0 statmod_1.5.0 bit64_4.6.0-1
#> [97] MASS_7.3-65